Ahrong Kim
AI-driven 3D Spatial Transcriptomics
Feb 25, 2025



Abstract:A comprehensive three-dimensional (3D) map of tissue architecture and gene expression is crucial for illuminating the complexity and heterogeneity of tissues across diverse biomedical applications. However, most spatial transcriptomics (ST) approaches remain limited to two-dimensional (2D) sections of tissue. Although current 3D ST methods hold promise, they typically require extensive tissue sectioning, are complex, are not compatible with non-destructive 3D tissue imaging technologies, and often lack scalability. Here, we present VOlumetrically Resolved Transcriptomics EXpression (VORTEX), an AI framework that leverages 3D tissue morphology and minimal 2D ST to predict volumetric 3D ST. By pretraining on diverse 3D morphology-transcriptomic pairs from heterogeneous tissue samples and then fine-tuning on minimal 2D ST data from a specific volume of interest, VORTEX learns both generic tissue-related and sample-specific morphological correlates of gene expression. This approach enables dense, high-throughput, and fast 3D ST, scaling seamlessly to large tissue volumes far beyond the reach of existing 3D ST techniques. By offering a cost-effective and minimally destructive route to obtaining volumetric molecular insights, we anticipate that VORTEX will accelerate biomarker discovery and our understanding of morphomolecular associations and cell states in complex tissues. Interactive 3D ST volumes can be viewed at https://vortex-demo.github.io/
Multimodal Whole Slide Foundation Model for Pathology
Nov 29, 2024



Abstract:The field of computational pathology has been transformed with recent advances in foundation models that encode histopathology region-of-interests (ROIs) into versatile and transferable feature representations via self-supervised learning (SSL). However, translating these advancements to address complex clinical challenges at the patient and slide level remains constrained by limited clinical data in disease-specific cohorts, especially for rare clinical conditions. We propose TITAN, a multimodal whole slide foundation model pretrained using 335,645 WSIs via visual self-supervised learning and vision-language alignment with corresponding pathology reports and 423,122 synthetic captions generated from a multimodal generative AI copilot for pathology. Without any finetuning or requiring clinical labels, TITAN can extract general-purpose slide representations and generate pathology reports that generalize to resource-limited clinical scenarios such as rare disease retrieval and cancer prognosis. We evaluate TITAN on diverse clinical tasks and find that TITAN outperforms both ROI and slide foundation models across machine learning settings such as linear probing, few-shot and zero-shot classification, rare cancer retrieval and cross-modal retrieval, and pathology report generation.
HEST-1k: A Dataset for Spatial Transcriptomics and Histology Image Analysis
Jun 23, 2024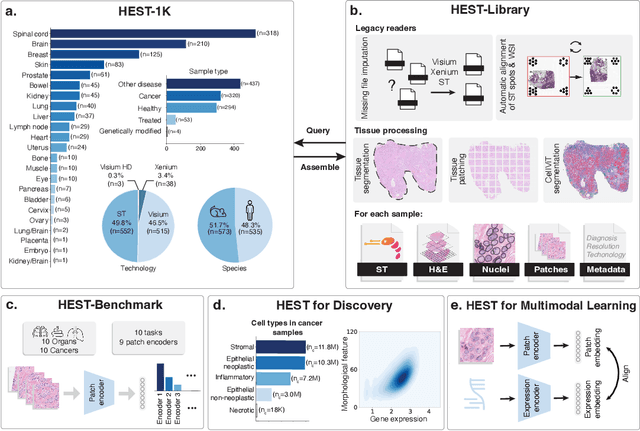
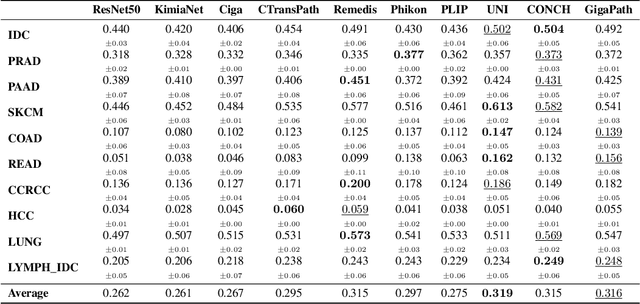
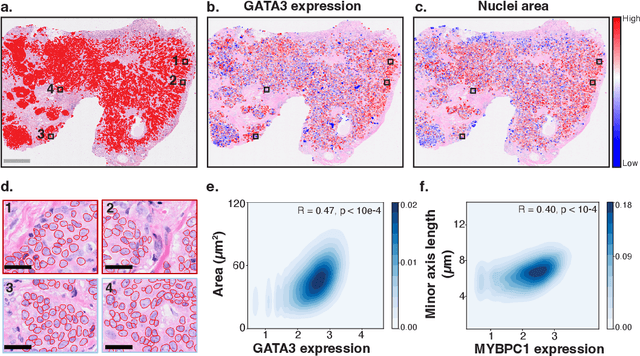
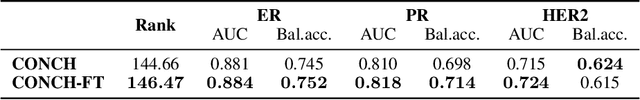
Abstract:Spatial transcriptomics (ST) enables interrogating the molecular composition of tissue with ever-increasing resolution, depth, and sensitivity. However, costs, rapidly evolving technology, and lack of standards have constrained computational methods in ST to narrow tasks and small cohorts. In addition, the underlying tissue morphology as reflected by H&E-stained whole slide images (WSIs) encodes rich information often overlooked in ST studies. Here, we introduce HEST-1k, a collection of 1,108 spatial transcriptomic profiles, each linked to a WSI and metadata. HEST-1k was assembled using HEST-Library from 131 public and internal cohorts encompassing 25 organs, two species (Homo Sapiens and Mus Musculus), and 320 cancer samples from 25 cancer types. HEST-1k processing enabled the identification of 1.5 million expression--morphology pairs and 60 million nuclei. HEST-1k is tested on three use cases: (1) benchmarking foundation models for histopathology (HEST-Benchmark), (2) biomarker identification, and (3) multimodal representation learning. HEST-1k, HEST-Library, and HEST-Benchmark can be freely accessed via https://github.com/mahmoodlab/hest.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge