Konstantin Hemker
AI-driven 3D Spatial Transcriptomics
Feb 25, 2025



Abstract:A comprehensive three-dimensional (3D) map of tissue architecture and gene expression is crucial for illuminating the complexity and heterogeneity of tissues across diverse biomedical applications. However, most spatial transcriptomics (ST) approaches remain limited to two-dimensional (2D) sections of tissue. Although current 3D ST methods hold promise, they typically require extensive tissue sectioning, are complex, are not compatible with non-destructive 3D tissue imaging technologies, and often lack scalability. Here, we present VOlumetrically Resolved Transcriptomics EXpression (VORTEX), an AI framework that leverages 3D tissue morphology and minimal 2D ST to predict volumetric 3D ST. By pretraining on diverse 3D morphology-transcriptomic pairs from heterogeneous tissue samples and then fine-tuning on minimal 2D ST data from a specific volume of interest, VORTEX learns both generic tissue-related and sample-specific morphological correlates of gene expression. This approach enables dense, high-throughput, and fast 3D ST, scaling seamlessly to large tissue volumes far beyond the reach of existing 3D ST techniques. By offering a cost-effective and minimally destructive route to obtaining volumetric molecular insights, we anticipate that VORTEX will accelerate biomarker discovery and our understanding of morphomolecular associations and cell states in complex tissues. Interactive 3D ST volumes can be viewed at https://vortex-demo.github.io/
Measuring Cross-Modal Interactions in Multimodal Models
Dec 20, 2024
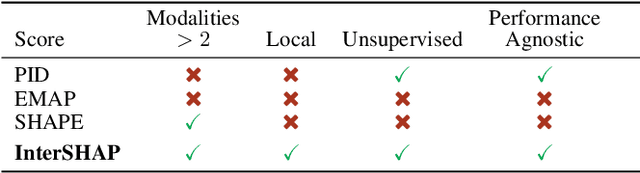
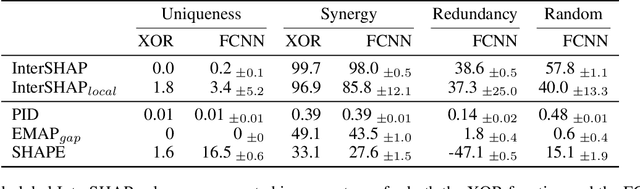
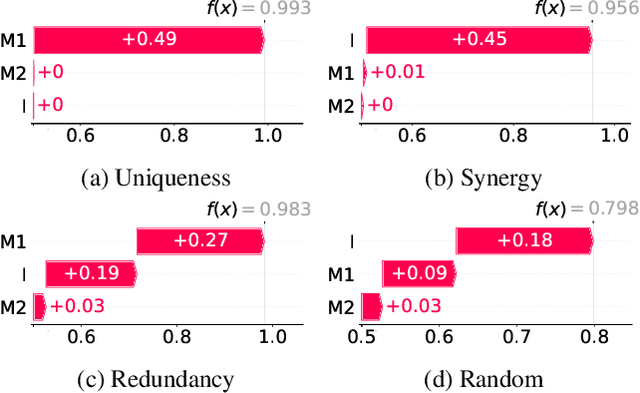
Abstract:Integrating AI in healthcare can greatly improve patient care and system efficiency. However, the lack of explainability in AI systems (XAI) hinders their clinical adoption, especially in multimodal settings that use increasingly complex model architectures. Most existing XAI methods focus on unimodal models, which fail to capture cross-modal interactions crucial for understanding the combined impact of multiple data sources. Existing methods for quantifying cross-modal interactions are limited to two modalities, rely on labelled data, and depend on model performance. This is problematic in healthcare, where XAI must handle multiple data sources and provide individualised explanations. This paper introduces InterSHAP, a cross-modal interaction score that addresses the limitations of existing approaches. InterSHAP uses the Shapley interaction index to precisely separate and quantify the contributions of the individual modalities and their interactions without approximations. By integrating an open-source implementation with the SHAP package, we enhance reproducibility and ease of use. We show that InterSHAP accurately measures the presence of cross-modal interactions, can handle multiple modalities, and provides detailed explanations at a local level for individual samples. Furthermore, we apply InterSHAP to multimodal medical datasets and demonstrate its applicability for individualised explanations.
PATHS: A Hierarchical Transformer for Efficient Whole Slide Image Analysis
Nov 27, 2024Abstract:Computational analysis of whole slide images (WSIs) has seen significant research progress in recent years, with applications ranging across important diagnostic and prognostic tasks such as survival or cancer subtype prediction. Many state-of-the-art models process the entire slide - which may be as large as $150,000 \times 150,000$ pixels - as a bag of many patches, the size of which necessitates computationally cheap feature aggregation methods. However, a large proportion of these patches are uninformative, such as those containing only healthy or adipose tissue, adding significant noise and size to the bag. We propose Pathology Transformer with Hierarchical Selection (PATHS), a novel top-down method for hierarchical weakly supervised representation learning on slide-level tasks in computational pathology. PATHS is inspired by the cross-magnification manner in which a human pathologist examines a slide, recursively filtering patches at each magnification level to a small subset relevant to the diagnosis. Our method overcomes the complications of processing the entire slide, enabling quadratic self-attention and providing a simple interpretable measure of region importance. We apply PATHS to five datasets of The Cancer Genome Atlas (TCGA), and achieve superior performance on slide-level prediction tasks when compared to previous methods, despite processing only a small proportion of the slide.
MM-Lego: Modular Biomedical Multimodal Models with Minimal Fine-Tuning
May 30, 2024



Abstract:Learning holistic computational representations in physical, chemical or biological systems requires the ability to process information from different distributions and modalities within the same model. Thus, the demand for multimodal machine learning models has sharply risen for modalities that go beyond vision and language, such as sequences, graphs, time series, or tabular data. While there are many available multimodal fusion and alignment approaches, most of them require end-to-end training, scale quadratically with the number of modalities, cannot handle cases of high modality imbalance in the training set, or are highly topology-specific, making them too restrictive for many biomedical learning tasks. This paper presents Multimodal Lego (MM-Lego), a modular and general-purpose fusion and model merging framework to turn any set of encoders into a competitive multimodal model with no or minimal fine-tuning. We achieve this by introducing a wrapper for unimodal encoders that enforces lightweight dimensionality assumptions between modalities and harmonises their representations by learning features in the frequency domain to enable model merging with little signal interference. We show that MM-Lego 1) can be used as a model merging method which achieves competitive performance with end-to-end fusion models without any fine-tuning, 2) can operate on any unimodal encoder, and 3) is a model fusion method that, with minimal fine-tuning, achieves state-of-the-art results on six benchmarked multimodal biomedical tasks.
HEALNet -- Hybrid Multi-Modal Fusion for Heterogeneous Biomedical Data
Nov 20, 2023



Abstract:Technological advances in medical data collection such as high-resolution histopathology and high-throughput genomic sequencing have contributed to the rising requirement for multi-modal biomedical modelling, specifically for image, tabular, and graph data. Most multi-modal deep learning approaches use modality-specific architectures that are trained separately and cannot capture the crucial cross-modal information that motivates the integration of different data sources. This paper presents the Hybrid Early-fusion Attention Learning Network (HEALNet): a flexible multi-modal fusion architecture, which a) preserves modality-specific structural information, b) captures the cross-modal interactions and structural information in a shared latent space, c) can effectively handle missing modalities during training and inference, and d) enables intuitive model inspection by learning on the raw data input instead of opaque embeddings. We conduct multi-modal survival analysis on Whole Slide Images and Multi-omic data on four cancer cohorts of The Cancer Genome Atlas (TCGA). HEALNet achieves state-of-the-art performance, substantially improving over both uni-modal and recent multi-modal baselines, whilst being robust in scenarios with missing modalities.
CGXplain: Rule-Based Deep Neural Network Explanations Using Dual Linear Programs
Apr 11, 2023


Abstract:Rule-based surrogate models are an effective and interpretable way to approximate a Deep Neural Network's (DNN) decision boundaries, allowing humans to easily understand deep learning models. Current state-of-the-art decompositional methods, which are those that consider the DNN's latent space to extract more exact rule sets, manage to derive rule sets at high accuracy. However, they a) do not guarantee that the surrogate model has learned from the same variables as the DNN (alignment), b) only allow to optimise for a single objective, such as accuracy, which can result in excessively large rule sets (complexity), and c) use decision tree algorithms as intermediate models, which can result in different explanations for the same DNN (stability). This paper introduces the CGX (Column Generation eXplainer) to address these limitations - a decompositional method using dual linear programming to extract rules from the hidden representations of the DNN. This approach allows to optimise for any number of objectives and empowers users to tweak the explanation model to their needs. We evaluate our results on a wide variety of tasks and show that CGX meets all three criteria, by having exact reproducibility of the explanation model that guarantees stability and reduces the rule set size by >80% (complexity) at equivalent or improved accuracy and fidelity across tasks (alignment).
Feature Synergy, Redundancy, and Independence in Global Model Explanations using SHAP Vector Decomposition
Jul 26, 2021


Abstract:We offer a new formalism for global explanations of pairwise feature dependencies and interactions in supervised models. Building upon SHAP values and SHAP interaction values, our approach decomposes feature contributions into synergistic, redundant and independent components (S-R-I decomposition of SHAP vectors). We propose a geometric interpretation of the components and formally prove its basic properties. Finally, we demonstrate the utility of synergy, redundancy and independence by applying them to a constructed data set and model.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge