Laura Wenderoth
Measuring Cross-Modal Interactions in Multimodal Models
Dec 20, 2024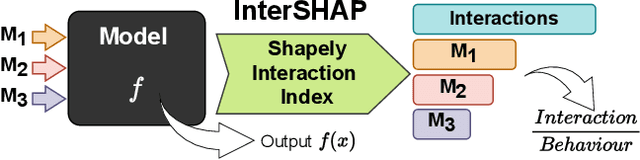
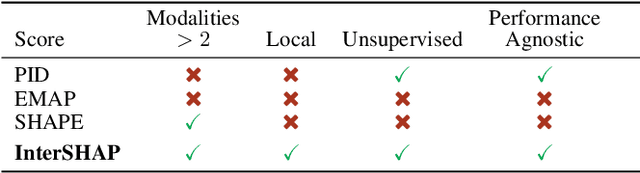
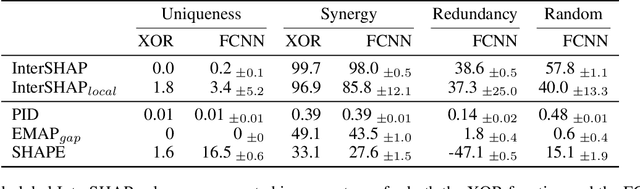
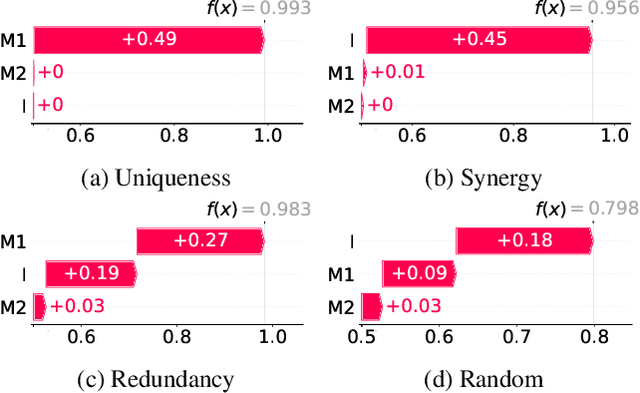
Abstract:Integrating AI in healthcare can greatly improve patient care and system efficiency. However, the lack of explainability in AI systems (XAI) hinders their clinical adoption, especially in multimodal settings that use increasingly complex model architectures. Most existing XAI methods focus on unimodal models, which fail to capture cross-modal interactions crucial for understanding the combined impact of multiple data sources. Existing methods for quantifying cross-modal interactions are limited to two modalities, rely on labelled data, and depend on model performance. This is problematic in healthcare, where XAI must handle multiple data sources and provide individualised explanations. This paper introduces InterSHAP, a cross-modal interaction score that addresses the limitations of existing approaches. InterSHAP uses the Shapley interaction index to precisely separate and quantify the contributions of the individual modalities and their interactions without approximations. By integrating an open-source implementation with the SHAP package, we enhance reproducibility and ease of use. We show that InterSHAP accurately measures the presence of cross-modal interactions, can handle multiple modalities, and provides detailed explanations at a local level for individual samples. Furthermore, we apply InterSHAP to multimodal medical datasets and demonstrate its applicability for individualised explanations.
Diagnosing Medical Datasets with Training Dynamics
Nov 03, 2024Abstract:This study explores the potential of using training dynamics as an automated alternative to human annotation for evaluating the quality of training data. The framework used is Data Maps, which classifies data points into categories such as easy-to-learn, hard-to-learn, and ambiguous (Swayamdipta et al., 2020). Swayamdipta et al. (2020) highlight that difficult-to-learn examples often contain errors, and ambiguous cases significantly impact model training. To confirm the reliability of these findings, we replicated the experiments using a challenging dataset, with a focus on medical question answering. In addition to text comprehension, this field requires the acquisition of detailed medical knowledge, which further complicates the task. A comprehensive evaluation was conducted to assess the feasibility and transferability of the Data Maps framework to the medical domain. The evaluation indicates that the framework is unsuitable for addressing datasets' unique challenges in answering medical questions.
Exploring Multi-Modality Dynamics: Insights and Challenges in Multimodal Fusion for Biomedical Tasks
Nov 01, 2024



Abstract:This paper investigates the MM dynamics approach proposed by Han et al. (2022) for multi-modal fusion in biomedical classification tasks. The MM dynamics algorithm integrates feature-level and modality-level informativeness to dynamically fuse modalities for improved classification performance. However, our analysis reveals several limitations and challenges in replicating and extending the results of MM dynamics. We found that feature informativeness improves performance and explainability, while modality informativeness does not provide significant advantages and can lead to performance degradation. Based on these results, we have extended feature informativeness to image data, resulting in the development of Image MM dynamics. Although this approach showed promising qualitative results, it did not outperform baseline methods quantitatively.
Self-supervision for medical image classification: state-of-the-art performance with ~100 labeled training samples per class
Apr 11, 2023Abstract:Is self-supervised deep learning (DL) for medical image analysis already a serious alternative to the de facto standard of end-to-end trained supervised DL? We tackle this question for medical image classification, with a particular focus on one of the currently most limiting factors of the field: the (non-)availability of labeled data. Based on three common medical imaging modalities (bone marrow microscopy, gastrointestinal endoscopy, dermoscopy) and publicly available data sets, we analyze the performance of self-supervised DL within the self-distillation with no labels (DINO) framework. After learning an image representation without use of image labels, conventional machine learning classifiers are applied. The classifiers are fit using a systematically varied number of labeled data (1-1000 samples per class). Exploiting the learned image representation, we achieve state-of-the-art classification performance for all three imaging modalities and data sets with only a fraction of between 1% and 10% of the available labeled data and about 100 labeled samples per class.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge