René Werner
Automating Parameter Selection in Deep Image Prior for Fluorescence Microscopy Image Denoising via Similarity-Based Parameter Transfer
Jan 17, 2026Abstract:Unsupervised deep image prior (DIP) addresses shortcomings of training data requirements and limited generalization associated with supervised deep learning. The performance of DIP depends on the network architecture and the stopping point of its iterative process. Optimizing these parameters for a new image requires time, restricting DIP application in domains where many images need to be processed. Focusing on fluorescence microscopy data, we hypothesize that similar images share comparable optimal parameter configurations for DIP-based denoising, potentially enabling optimization-free DIP for fluorescence microscopy. We generated a calibration (n=110) and validation set (n=55) of semantically different images from an open-source dataset for a network architecture search targeted towards ideal U-net architectures and stopping points. The calibration set represented our transfer basis. The validation set enabled the assessment of which image similarity criterion yields the best results. We then implemented AUTO-DIP, a pipeline for automatic parameter transfer, and compared it to the originally published DIP configuration (baseline) and a state-of-the-art image-specific variational denoising approach. We show that a parameter transfer from the calibration dataset to a test image based on only image metadata similarity (e.g., microscope type, imaged specimen) leads to similar and better performance than a transfer based on quantitative image similarity measures. AUTO-DIP outperforms the baseline DIP (DIP with original DIP parameters) as well as the variational denoising approaches for several open-source test datasets of varying complexity, particularly for very noisy inputs. Applications to locally acquired fluorescence microscopy images further proved superiority of AUTO-DIP.
Oriented histogram-based vector field embedding for characterizing 4D CT data sets in radiotherapy
Nov 25, 2024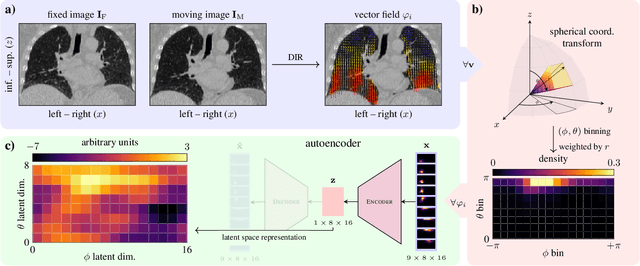
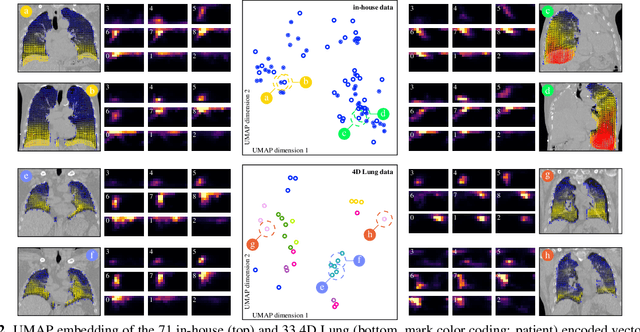
Abstract:In lung radiotherapy, the primary objective is to optimize treatment outcomes by minimizing exposure to healthy tissues while delivering the prescribed dose to the target volume. The challenge lies in accounting for lung tissue motion due to breathing, which impacts precise treatment alignment. To address this, the paper proposes a prospective approach that relies solely on pre-treatment information, such as planning CT scans and derived data like vector fields from deformable image registration. This data is compared to analogous patient data to tailor treatment strategies, i.e., to be able to review treatment parameters and success for similar patients. To allow for such a comparison, an embedding and clustering strategy of prospective patient data is needed. Therefore, the main focus of this study lies on reducing the dimensionality of deformable registration-based vector fields by employing a voxel-wise spherical coordinate transformation and a low-dimensional 2D oriented histogram representation. Afterwards, a fully unsupervised UMAP embedding of the encoded vector fields (i.e., patient-specific motion information) becomes applicable. The functionality of the proposed method is demonstrated with 71 in-house acquired 4D CT data sets and 33 external 4D CT data sets. A comprehensive analysis of the patient clusters is conducted, focusing on the similarity of breathing patterns of clustered patients. The proposed general approach of reducing the dimensionality of registration vector fields by encoding the inherent information into oriented histograms is, however, applicable to other tasks.
Cluster-based human-in-the-loop strategy for improving machine learning-based circulating tumor cell detection in liquid biopsy
Nov 25, 2024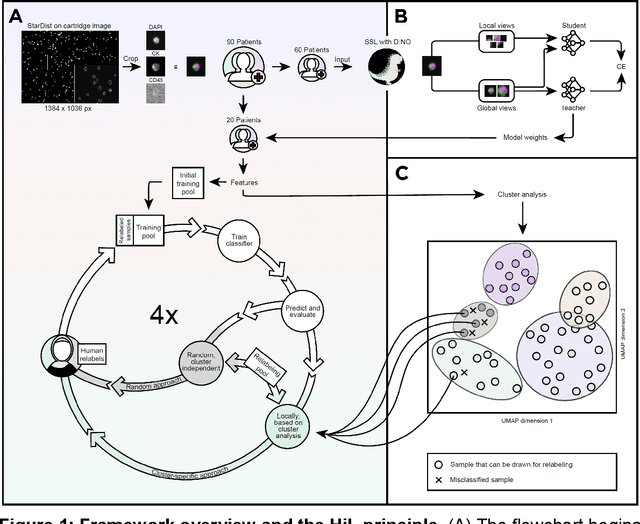



Abstract:Detection and differentiation of circulating tumor cells (CTCs) and non-CTCs in blood draws of cancer patients pose multiple challenges. While the gold standard relies on tedious manual evaluation of an automatically generated selection of images, machine learning (ML) techniques offer the potential to automate these processes. However, human assessment remains indispensable when the ML system arrives at uncertain or wrong decisions due to an insufficient set of labeled training data. This study introduces a human-in-the-loop (HiL) strategy for improving ML-based CTC detection. We combine self-supervised deep learning and a conventional ML-based classifier and propose iterative targeted sampling and labeling of new unlabeled training samples by human experts. The sampling strategy is based on the classification performance of local latent space clusters. The advantages of the proposed approach compared to naive random sampling are demonstrated for liquid biopsy data from patients with metastatic breast cancer.
Surgical tool classification and localization: results and methods from the MICCAI 2022 SurgToolLoc challenge
May 11, 2023



Abstract:The ability to automatically detect and track surgical instruments in endoscopic videos can enable transformational interventions. Assessing surgical performance and efficiency, identifying skilled tool use and choreography, and planning operational and logistical aspects of OR resources are just a few of the applications that could benefit. Unfortunately, obtaining the annotations needed to train machine learning models to identify and localize surgical tools is a difficult task. Annotating bounding boxes frame-by-frame is tedious and time-consuming, yet large amounts of data with a wide variety of surgical tools and surgeries must be captured for robust training. Moreover, ongoing annotator training is needed to stay up to date with surgical instrument innovation. In robotic-assisted surgery, however, potentially informative data like timestamps of instrument installation and removal can be programmatically harvested. The ability to rely on tool installation data alone would significantly reduce the workload to train robust tool-tracking models. With this motivation in mind we invited the surgical data science community to participate in the challenge, SurgToolLoc 2022. The goal was to leverage tool presence data as weak labels for machine learning models trained to detect tools and localize them in video frames with bounding boxes. We present the results of this challenge along with many of the team's efforts. We conclude by discussing these results in the broader context of machine learning and surgical data science. The training data used for this challenge consisting of 24,695 video clips with tool presence labels is also being released publicly and can be accessed at https://console.cloud.google.com/storage/browser/isi-surgtoolloc-2022.
Self-supervision for medical image classification: state-of-the-art performance with ~100 labeled training samples per class
Apr 11, 2023Abstract:Is self-supervised deep learning (DL) for medical image analysis already a serious alternative to the de facto standard of end-to-end trained supervised DL? We tackle this question for medical image classification, with a particular focus on one of the currently most limiting factors of the field: the (non-)availability of labeled data. Based on three common medical imaging modalities (bone marrow microscopy, gastrointestinal endoscopy, dermoscopy) and publicly available data sets, we analyze the performance of self-supervised DL within the self-distillation with no labels (DINO) framework. After learning an image representation without use of image labels, conventional machine learning classifiers are applied. The classifiers are fit using a systematically varied number of labeled data (1-1000 samples per class). Exploiting the learned image representation, we achieve state-of-the-art classification performance for all three imaging modalities and data sets with only a fraction of between 1% and 10% of the available labeled data and about 100 labeled samples per class.
Skin Lesion Classification Using Ensembles of Multi-Resolution EfficientNets with Meta Data
Oct 09, 2019
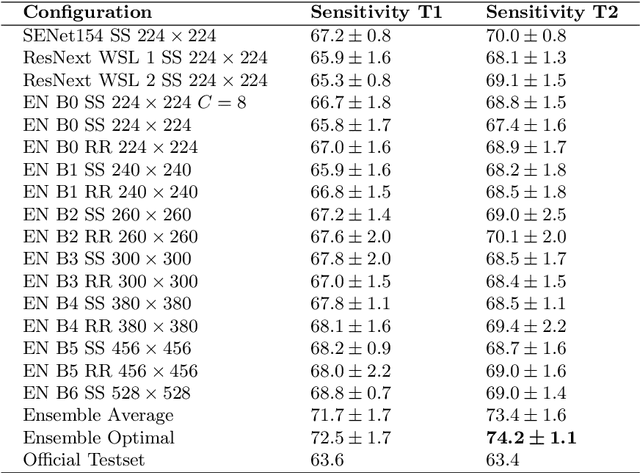
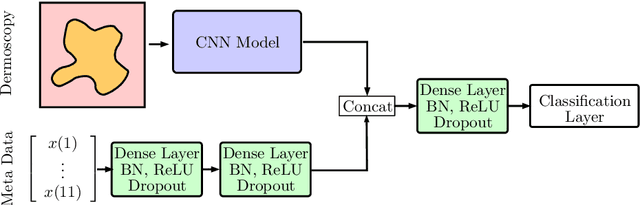
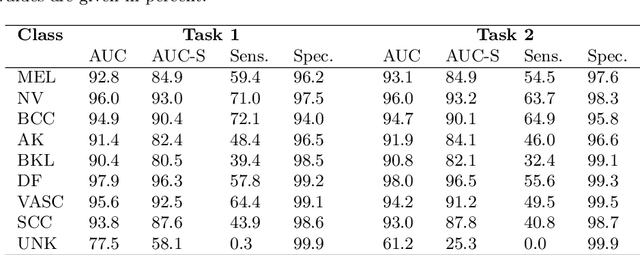
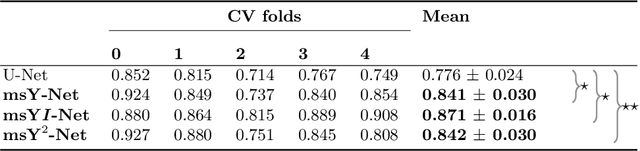
Abstract:In this paper, we describe our method for the ISIC 2019 Skin Lesion Classification Challenge. The challenge comes with two tasks. For task 1, skin lesions have to be classified based on dermoscopic images. For task 2, dermoscopic images and additional patient meta data have to be used. A diverse dataset of 25000 images was provided for training, containing images from eight classes. The final test set contains an additional, unknown class. We address this challenging problem with a simple, data driven approach by including external data with skin lesions types that are not present in the training set. Furthermore, multi-class skin lesion classification comes with the problem of severe class imbalance. We try to overcome this problem by using loss balancing. Also, the dataset contains images with very different resolutions. We take care of this property by considering different model input resolutions and different cropping strategies. To incorporate meta data such as age, anatomical site, and sex, we use an additional dense neural network and fuse its features with the CNN. We aggregate all our models with an ensembling strategy where we search for the optimal subset of models. Our best ensemble achieves a balanced accuracy of 74.2% using five-fold cross-validation. On the official test set our method is ranked first for both tasks with a balanced accuracy of 63.6% for task 1 and 63.4% for task 2.
Multi-scale fully convolutional neural networks for histopathology image segmentation: from nuclear aberrations to the global tissue architecture
Sep 24, 2019
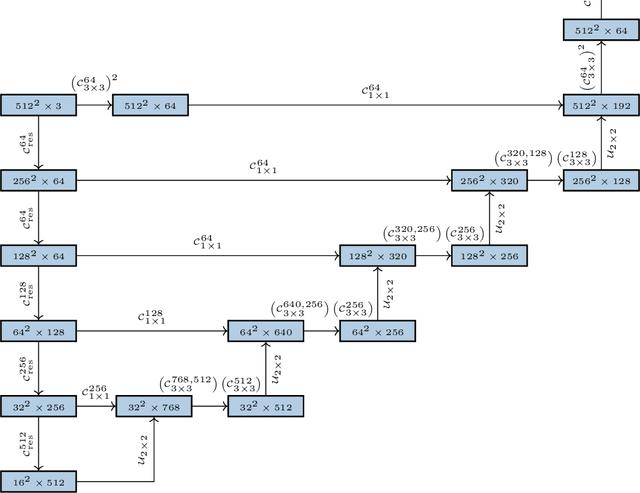
Abstract:Histopathologic diagnosis is dependent on simultaneous information from a broad range of scales, ranging from nuclear aberrations ($\approx \mathcal{O}(0.1 \mu m)$) over cellular structures ($\approx \mathcal{O}(10\mu m)$) to the global tissue architecture ($\gtrapprox \mathcal{O}(1 mm)$). Bearing in mind which information is employed by human pathologists, we introduce and examine different strategies for the integration of multiple and widely separate spatial scales into common U-Net-based architectures. Based on this, we present a family of new, end-to-end trainable, multi-scale multi-encoder fully-convolutional neural networks for human modus operandi-inspired computer vision in histopathology.
Skin Lesion Classification Using CNNs with Patch-Based Attention and Diagnosis-Guided Loss Weighting
May 09, 2019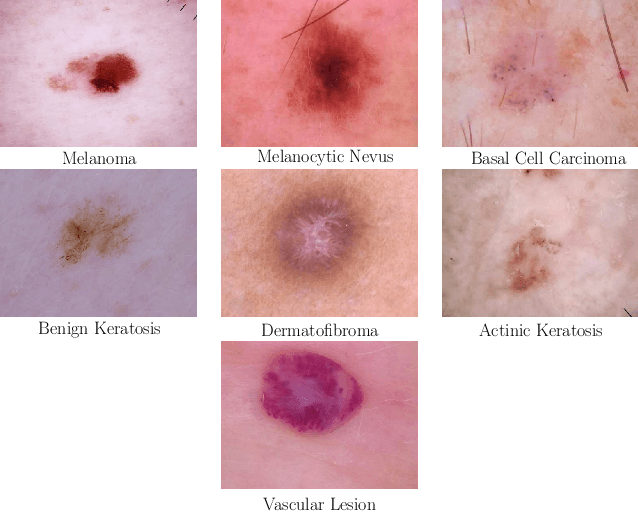
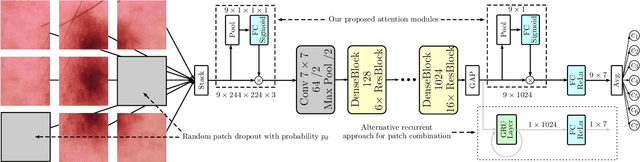
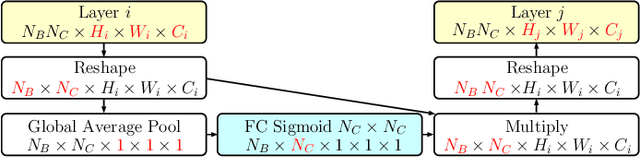
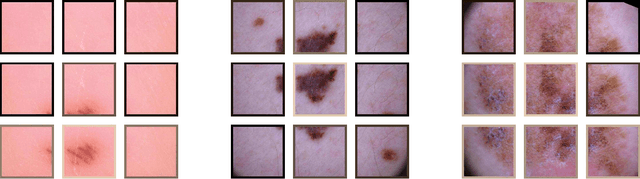
Abstract:Objective: This work addresses two key problems of skin lesion classification. The first problem is the effective use of high-resolution images with pretrained standard architectures for image classification. The second problem is the high class imbalance encountered in real-world multi-class datasets. Methods: To use high-resolution images, we propose a novel patch-based attention architecture that provides global context between small, high-resolution patches. We modify three pretrained architectures and study the performance of patch-based attention. To counter class imbalance problems, we compare oversampling, balanced batch sampling, and class-specific loss weighting. Additionally, we propose a novel diagnosis-guided loss weighting method which takes the method used for ground-truth annotation into account. Results: Our patch-based attention mechanism outperforms previous methods and improves the mean sensitivity by 7%. Class balancing significantly improves the mean sensitivity and we show that our diagnosis-guided loss weighting method improves the mean sensitivity by 3% over normal loss balancing. Conclusion: The novel patch-based attention mechanism can be integrated into pretrained architectures and provides global context between local patches while outperforming other patch-based methods. Hence, pretrained architectures can be readily used with high-resolution images without downsampling. The new diagnosis-guided loss weighting method outperforms other methods and allows for effective training when facing class imbalance. Significance: The proposed methods improve automatic skin lesion classification. They can be extended to other clinical applications where high-resolution image data and class imbalance are relevant.
3d-SMRnet: Achieving a new quality of MPI system matrix recovery by deep learning
May 08, 2019
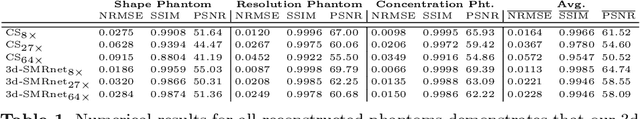
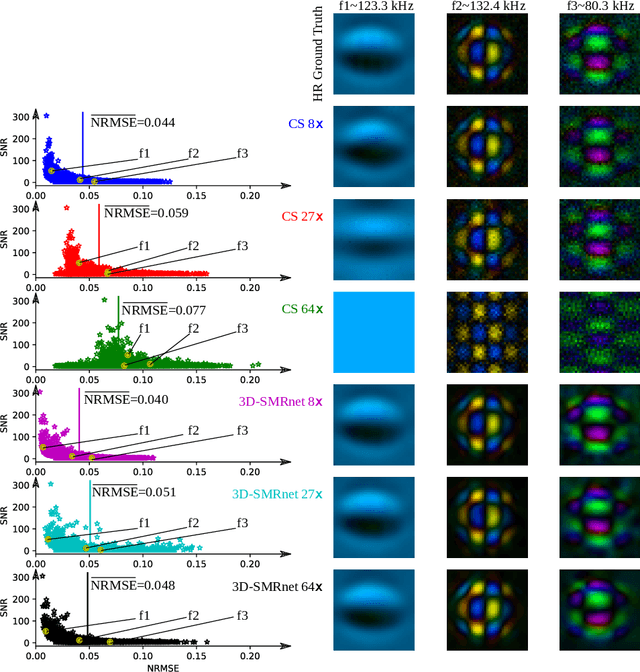
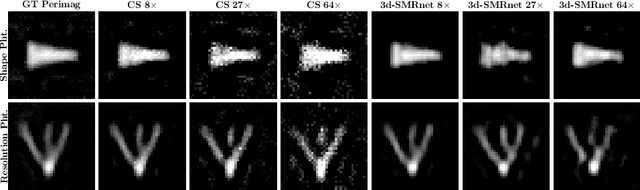
Abstract:Magnetic particle imaging (MPI) data is commonly reconstructed using a system matrix acquired in a time-consuming calibration measurement. The calibration approach has the important advantage over model-based reconstruction that it takes the complex particle physics as well as system imperfections into account. This benefit comes for the cost that the system matrix needs to be re-calibrated whenever the scan parameters, particle types or even the particle environment (e.g. viscosity or temperature) changes. One route for reducing the calibration time is the sampling of the system matrix at a subset of the spatial positions of the intended field-of-view and employing system matrix recovery. Recent approaches used compressed sensing (CS) and achieved subsampling factors up to 28 that still allowed reconstructing MPI images of sufficient quality. In this work, we propose a novel framework with a 3d-System Matrix Recovery Network and demonstrate it to recover a 3d system matrix with a subsampling factor of 64 in less than one minute and to outperform CS in terms of system matrix quality, reconstructed image quality, and processing time. The advantage of our method is demonstrated by reconstructing open access MPI datasets. The model is further shown to be capable of inferring system matrices for different particle types.
Skin Lesion Diagnosis using Ensembles, Unscaled Multi-Crop Evaluation and Loss Weighting
Aug 05, 2018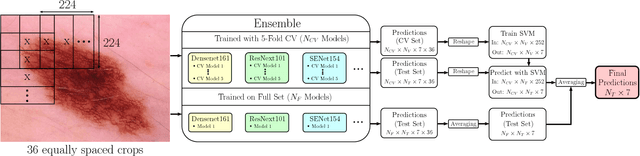
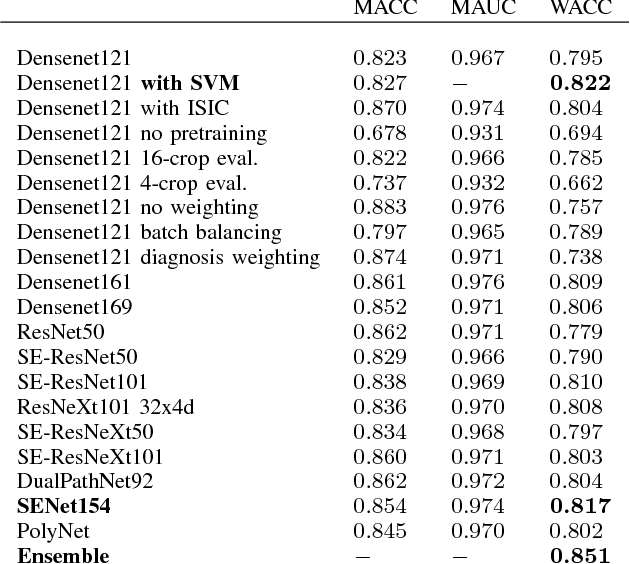
Abstract:In this paper we present the methods of our submission to the ISIC 2018 challenge for skin lesion diagnosis (Task 3). The dataset consists of 10000 images with seven image-level classes to be distinguished by an automated algorithm. We employ an ensemble of convolutional neural networks for this task. In particular, we fine-tune pretrained state-of-the-art deep learning models such as Densenet, SENet and ResNeXt. We identify heavy class imbalance as a key problem for this challenge and consider multiple balancing approaches such as loss weighting and balanced batch sampling. Another important feature of our pipeline is the use of a vast amount of unscaled crops for evaluation. Last, we consider meta learning approaches for the final predictions. Our team placed second at the challenge while being the best approach using only publicly available data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge