Nils Gessert
Multiple Sclerosis Lesion Activity Segmentation with Attention-Guided Two-Path CNNs
Aug 05, 2020



Abstract:Multiple sclerosis is an inflammatory autoimmune demyelinating disease that is characterized by lesions in the central nervous system. Typically, magnetic resonance imaging (MRI) is used for tracking disease progression. Automatic image processing methods can be used to segment lesions and derive quantitative lesion parameters. So far, methods have focused on lesion segmentation for individual MRI scans. However, for monitoring disease progression, \textit{lesion activity} in terms of new and enlarging lesions between two time points is a crucial biomarker. For this problem, several classic methods have been proposed, e.g., using difference volumes. Despite their success for single-volume lesion segmentation, deep learning approaches are still rare for lesion activity segmentation. In this work, convolutional neural networks (CNNs) are studied for lesion activity segmentation from two time points. For this task, CNNs are designed and evaluated that combine the information from two points in different ways. In particular, two-path architectures with attention-guided interactions are proposed that enable effective information exchange between the two time point's processing paths. It is demonstrated that deep learning-based methods outperform classic approaches and it is shown that attention-guided interactions significantly improve performance. Furthermore, the attention modules produce plausible attention maps that have a masking effect that suppresses old, irrelevant lesions. A lesion-wise false positive rate of 26.4% is achieved at a true positive rate of 74.2%, which is not significantly different from the interrater performance.
4D Spatio-Temporal Convolutional Networks for Object Position Estimation in OCT Volumes
Jul 02, 2020


Abstract:Tracking and localizing objects is a central problem in computer-assisted surgery. Optical coherence tomography (OCT) can be employed as an optical tracking system, due to its high spatial and temporal resolution. Recently, 3D convolutional neural networks (CNNs) have shown promising performance for pose estimation of a marker object using single volumetric OCT images. While this approach relied on spatial information only, OCT allows for a temporal stream of OCT image volumes capturing the motion of an object at high volumes rates. In this work, we systematically extend 3D CNNs to 4D spatio-temporal CNNs to evaluate the impact of additional temporal information for marker object tracking. Across various architectures, our results demonstrate that using a stream of OCT volumes and employing 4D spatio-temporal convolutions leads to a 30% lower mean absolute error compared to single volume processing with 3D CNNs.
Spectral-Spatial Recurrent-Convolutional Networks for In-Vivo Hyperspectral Tumor Type Classification
Jul 02, 2020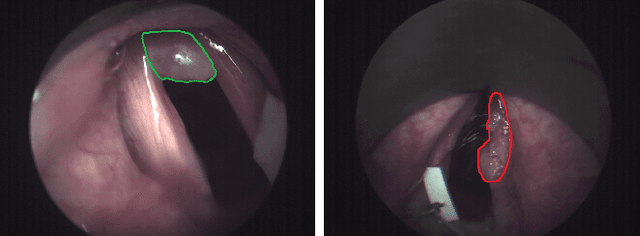
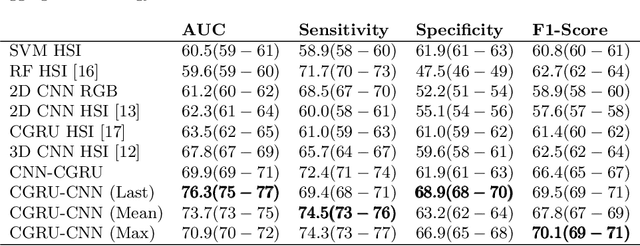
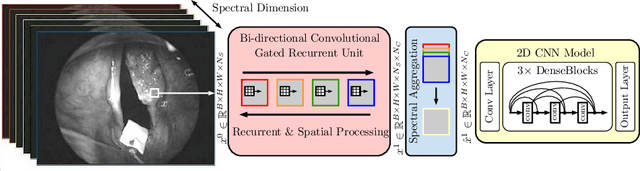
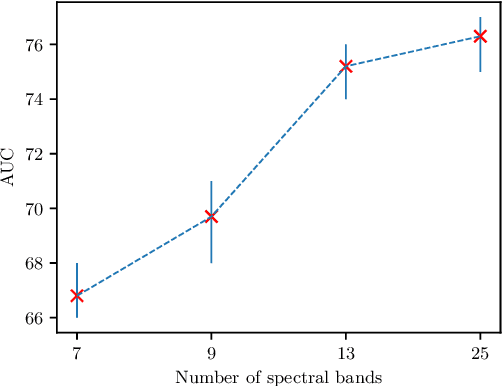
Abstract:Early detection of cancerous tissue is crucial for long-term patient survival. In the head and neck region, a typical diagnostic procedure is an endoscopic intervention where a medical expert manually assesses tissue using RGB camera images. While healthy and tumor regions are generally easier to distinguish, differentiating benign and malignant tumors is very challenging. This requires an invasive biopsy, followed by histological evaluation for diagnosis. Also, during tumor resection, tumor margins need to be verified by histological analysis. To avoid unnecessary tissue resection, a non-invasive, image-based diagnostic tool would be very valuable. Recently, hyperspectral imaging paired with deep learning has been proposed for this task, demonstrating promising results on ex-vivo specimens. In this work, we demonstrate the feasibility of in-vivo tumor type classification using hyperspectral imaging and deep learning. We analyze the value of using multiple hyperspectral bands compared to conventional RGB images and we study several machine learning models' ability to make use of the additional spectral information. Based on our insights, we address spectral and spatial processing using recurrent-convolutional models for effective spectral aggregating and spatial feature learning. Our best model achieves an AUC of 76.3%, significantly outperforming previous conventional and deep learning methods.
Deep learning with 4D spatio-temporal data representations for OCT-based force estimation
May 20, 2020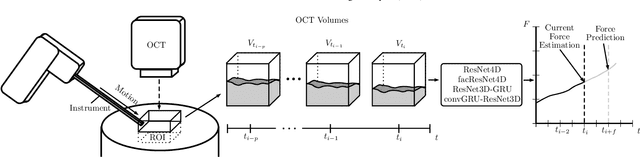
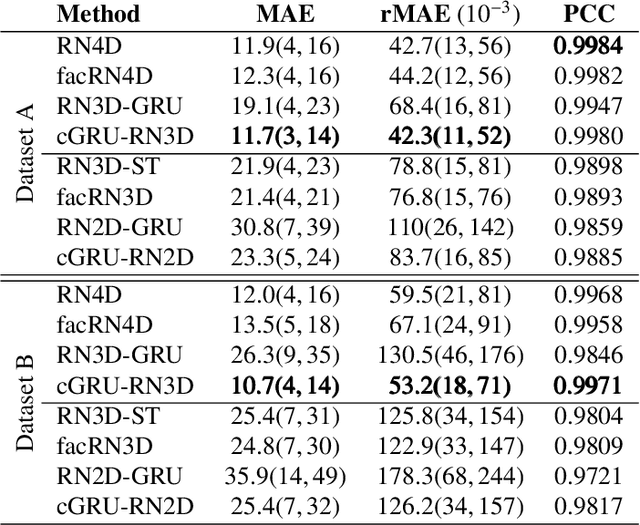
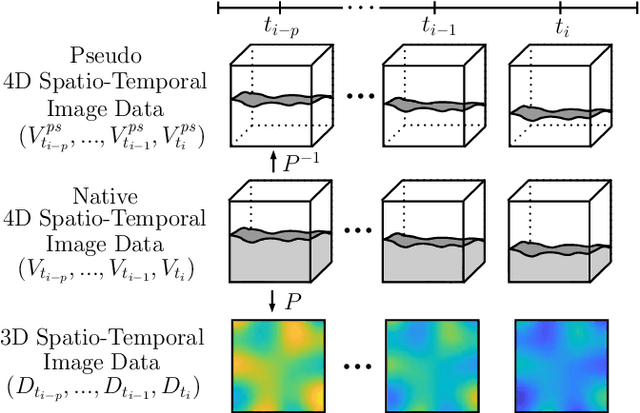
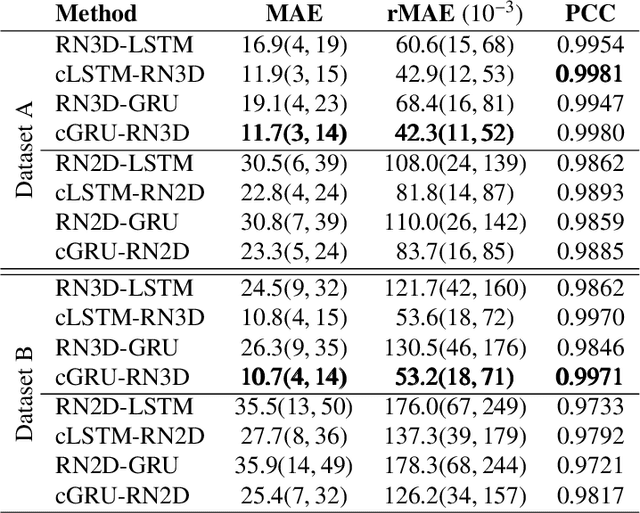
Abstract:Estimating the forces acting between instruments and tissue is a challenging problem for robot-assisted minimally-invasive surgery. Recently, numerous vision-based methods have been proposed to replace electro-mechanical approaches. Moreover, optical coherence tomography (OCT) and deep learning have been used for estimating forces based on deformation observed in volumetric image data. The method demonstrated the advantage of deep learning with 3D volumetric data over 2D depth images for force estimation. In this work, we extend the problem of deep learning-based force estimation to 4D spatio-temporal data with streams of 3D OCT volumes. For this purpose, we design and evaluate several methods extending spatio-temporal deep learning to 4D which is largely unexplored so far. Furthermore, we provide an in-depth analysis of multi-dimensional image data representations for force estimation, comparing our 4D approach to previous, lower-dimensional methods. Also, we analyze the effect of temporal information and we study the prediction of short-term future force values, which could facilitate safety features. For our 4D force estimation architectures, we find that efficient decoupling of spatial and temporal processing is advantageous. We show that using 4D spatio-temporal data outperforms all previously used data representations with a mean absolute error of 10.7mN. We find that temporal information is valuable for force estimation and we demonstrate the feasibility of force prediction.
4D Spatio-Temporal Deep Learning with 4D fMRI Data for Autism Spectrum Disorder Classification
Apr 21, 2020

Abstract:Autism spectrum disorder (ASD) is associated with behavioral and communication problems. Often, functional magnetic resonance imaging (fMRI) is used to detect and characterize brain changes related to the disorder. Recently, machine learning methods have been employed to reveal new patterns by trying to classify ASD from spatio-temporal fMRI images. Typically, these methods have either focused on temporal or spatial information processing. Instead, we propose a 4D spatio-temporal deep learning approach for ASD classification where we jointly learn from spatial and temporal data. We employ 4D convolutional neural networks and convolutional-recurrent models which outperform a previous approach with an F1-score of 0.71 compared to an F1-score of 0.65.
Spatio-spectral deep learning methods for in-vivo hyperspectral laryngeal cancer detection
Apr 21, 2020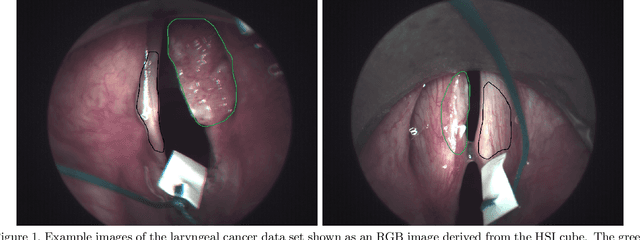
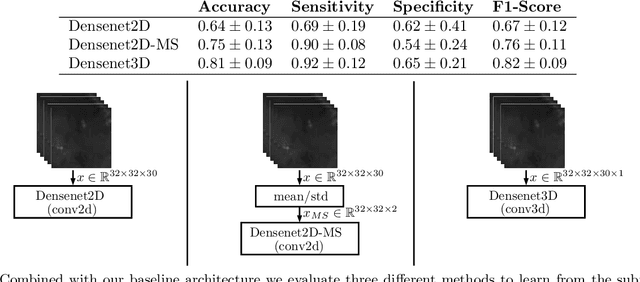
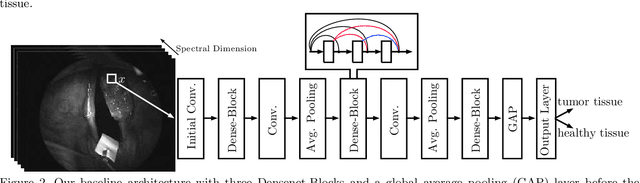
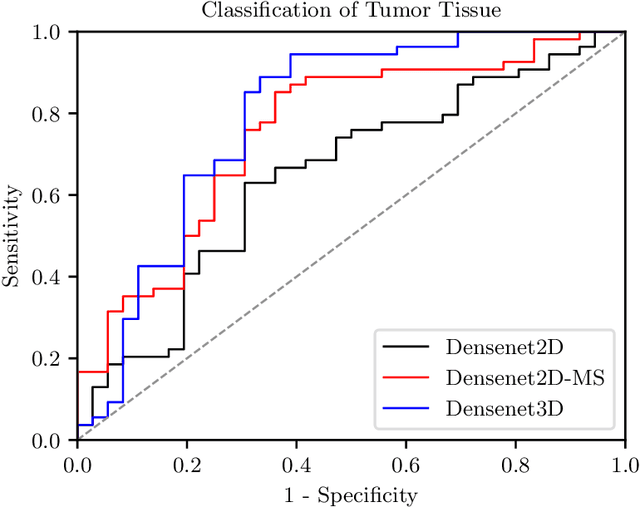
Abstract:Early detection of head and neck tumors is crucial for patient survival. Often, diagnoses are made based on endoscopic examination of the larynx followed by biopsy and histological analysis, leading to a high inter-observer variability due to subjective assessment. In this regard, early non-invasive diagnostics independent of the clinician would be a valuable tool. A recent study has shown that hyperspectral imaging (HSI) can be used for non-invasive detection of head and neck tumors, as precancerous or cancerous lesions show specific spectral signatures that distinguish them from healthy tissue. However, HSI data processing is challenging due to high spectral variations, various image interferences, and the high dimensionality of the data. Therefore, performance of automatic HSI analysis has been limited and so far, mostly ex-vivo studies have been presented with deep learning. In this work, we analyze deep learning techniques for in-vivo hyperspectral laryngeal cancer detection. For this purpose we design and evaluate convolutional neural networks (CNNs) with 2D spatial or 3D spatio-spectral convolutions combined with a state-of-the-art Densenet architecture. For evaluation, we use an in-vivo data set with HSI of the oral cavity or oropharynx. Overall, we present multiple deep learning techniques for in-vivo laryngeal cancer detection based on HSI and we show that jointly learning from the spatial and spectral domain improves classification accuracy notably. Our 3D spatio-spectral Densenet achieves an average accuracy of 81%.
A Deep Learning Approach for Motion Forecasting Using 4D OCT Data
Apr 21, 2020

Abstract:Forecasting motion of a specific target object is a common problem for surgical interventions, e.g. for localization of a target region, guidance for surgical interventions, or motion compensation. Optical coherence tomography (OCT) is an imaging modality with a high spatial and temporal resolution. Recently, deep learning methods have shown promising performance for OCT-based motion estimation based on two volumetric images. We extend this approach and investigate whether using a time series of volumes enables motion forecasting. We propose 4D spatio-temporal deep learning for end-to-end motion forecasting and estimation using a stream of OCT volumes. We design and evaluate five different 3D and 4D deep learning methods using a tissue data set. Our best performing 4D method achieves motion forecasting with an overall average correlation coefficient of 97.41%, while also improving motion estimation performance by a factor of 2.5 compared to a previous 3D approach.
Spatio-Temporal Deep Learning Methods for Motion Estimation Using 4D OCT Image Data
Apr 21, 2020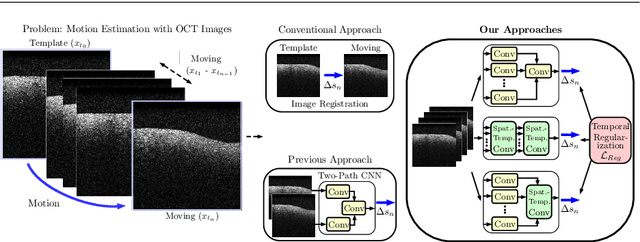
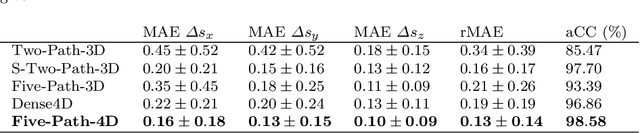
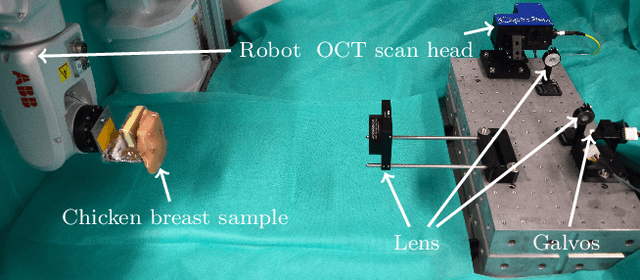
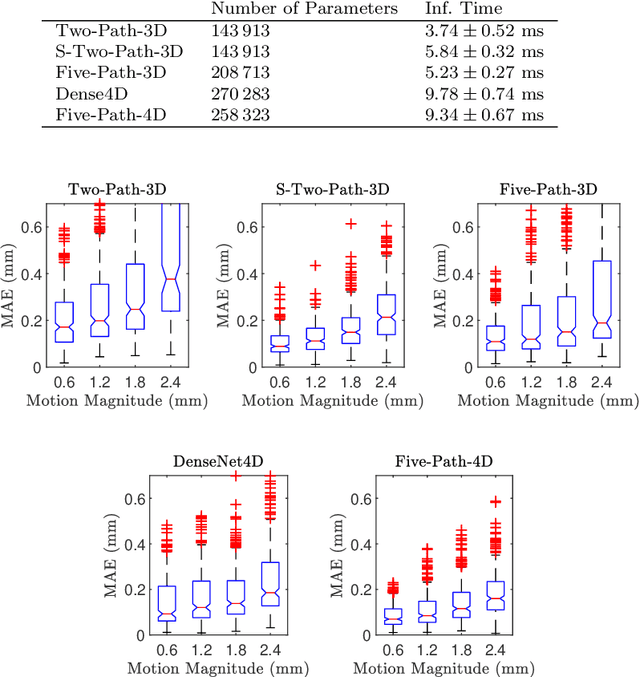
Abstract:Purpose. Localizing structures and estimating the motion of a specific target region are common problems for navigation during surgical interventions. Optical coherence tomography (OCT) is an imaging modality with a high spatial and temporal resolution that has been used for intraoperative imaging and also for motion estimation, for example, in the context of ophthalmic surgery or cochleostomy. Recently, motion estimation between a template and a moving OCT image has been studied with deep learning methods to overcome the shortcomings of conventional, feature-based methods. Methods. We investigate whether using a temporal stream of OCT image volumes can improve deep learning-based motion estimation performance. For this purpose, we design and evaluate several 3D and 4D deep learning methods and we propose a new deep learning approach. Also, we propose a temporal regularization strategy at the model output. Results. Using a tissue dataset without additional markers, our deep learning methods using 4D data outperform previous approaches. The best performing 4D architecture achieves an correlation coefficient (aCC) of 98.58% compared to 85.0% of a previous 3D deep learning method. Also, our temporal regularization strategy at the output further improves 4D model performance to an aCC of 99.06%. In particular, our 4D method works well for larger motion and is robust towards image rotations and motion distortions. Conclusions. We propose 4D spatio-temporal deep learning for OCT-based motion estimation. On a tissue dataset, we find that using 4D information for the model input improves performance while maintaining reasonable inference times. Our regularization strategy demonstrates that additional temporal information is also beneficial at the model output.
4D Deep Learning for Multiple Sclerosis Lesion Activity Segmentation
Apr 20, 2020

Abstract:Multiple sclerosis lesion activity segmentation is the task of detecting new and enlarging lesions that appeared between a baseline and a follow-up brain MRI scan. While deep learning methods for single-scan lesion segmentation are common, deep learning approaches for lesion activity have only been proposed recently. Here, a two-path architecture processes two 3D MRI volumes from two time points. In this work, we investigate whether extending this problem to full 4D deep learning using a history of MRI volumes and thus an extended baseline can improve performance. For this purpose, we design a recurrent multi-encoder-decoder architecture for processing 4D data. We find that adding more temporal information is beneficial and our proposed architecture outperforms previous approaches with a lesion-wise true positive rate of 0.84 at a lesion-wise false positive rate of 0.19.
Learning Preference-Based Similarities from Face Images using Siamese Multi-Task CNNs
Jan 25, 2020



Abstract:Online dating has become a common occurrence over the last few decades. A key challenge for online dating platforms is to determine suitable matches for their users. A lot of dating services rely on self-reported user traits and preferences for matching. At the same time, some services largely rely on user images and thus initial visual preference. Especially for the latter approach, previous research has attempted to capture users' visual preferences for automatic match recommendation. These approaches are mostly based on the assumption that physical attraction is the key factor for relationship formation and personal preferences, interests, and attitude are largely neglected. Deep learning approaches have shown that a variety of properties can be predicted from human faces to some degree, including age, health and even personality traits. Therefore, we investigate the feasibility of bridging image-based matching and matching with personal interests, preferences, and attitude. We approach the problem in a supervised manner by predicting similarity scores between two users based on images of their faces only. The ground-truth for the similarity matching scores is determined by a test that aims to capture users' preferences, interests, and attitude that are relevant for forming romantic relationships. The images are processed by a Siamese Multi-Task deep learning architecture. We find a statistically significant correlation between predicted and target similarity scores. Thus, our results indicate that learning similarities in terms of interests, preferences, and attitude from face images appears to be feasible to some degree.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge