Frederic Madesta
Beyond the LUMIR challenge: The pathway to foundational registration models
May 30, 2025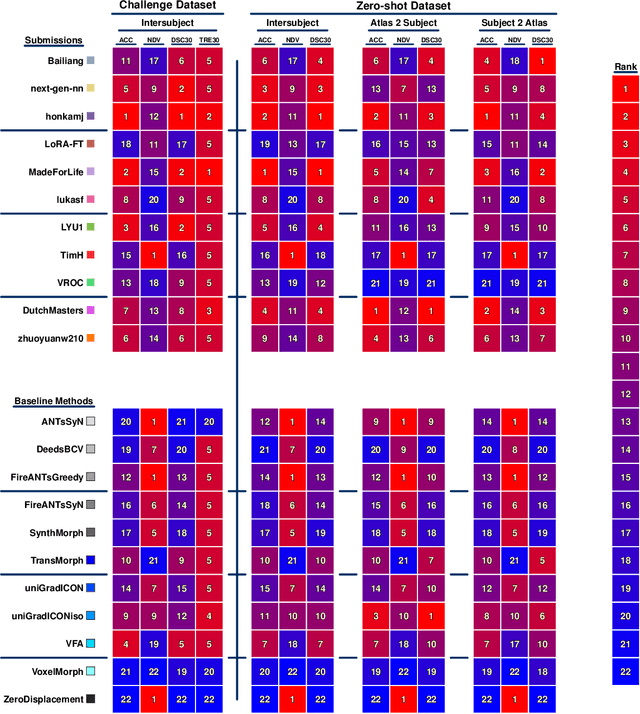
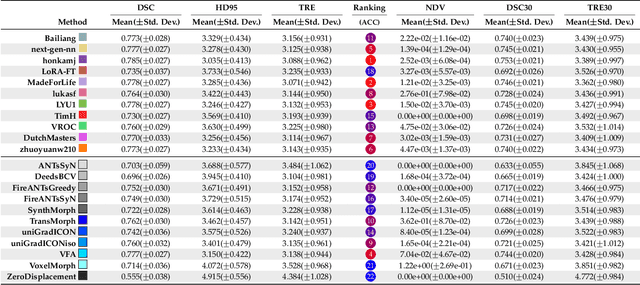
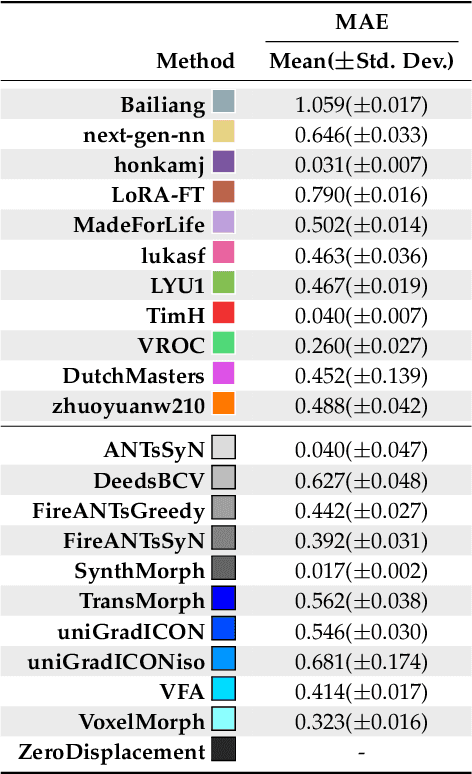
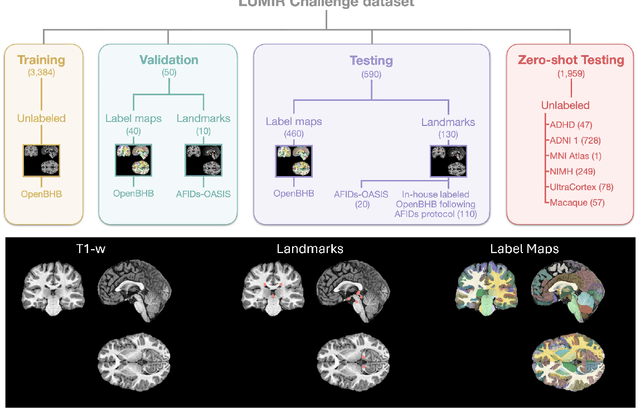
Abstract:Medical image challenges have played a transformative role in advancing the field, catalyzing algorithmic innovation and establishing new performance standards across diverse clinical applications. Image registration, a foundational task in neuroimaging pipelines, has similarly benefited from the Learn2Reg initiative. Building on this foundation, we introduce the Large-scale Unsupervised Brain MRI Image Registration (LUMIR) challenge, a next-generation benchmark designed to assess and advance unsupervised brain MRI registration. Distinct from prior challenges that leveraged anatomical label maps for supervision, LUMIR removes this dependency by providing over 4,000 preprocessed T1-weighted brain MRIs for training without any label maps, encouraging biologically plausible deformation modeling through self-supervision. In addition to evaluating performance on 590 held-out test subjects, LUMIR introduces a rigorous suite of zero-shot generalization tasks, spanning out-of-domain imaging modalities (e.g., FLAIR, T2-weighted, T2*-weighted), disease populations (e.g., Alzheimer's disease), acquisition protocols (e.g., 9.4T MRI), and species (e.g., macaque brains). A total of 1,158 subjects and over 4,000 image pairs were included for evaluation. Performance was assessed using both segmentation-based metrics (Dice coefficient, 95th percentile Hausdorff distance) and landmark-based registration accuracy (target registration error). Across both in-domain and zero-shot tasks, deep learning-based methods consistently achieved state-of-the-art accuracy while producing anatomically plausible deformation fields. The top-performing deep learning-based models demonstrated diffeomorphic properties and inverse consistency, outperforming several leading optimization-based methods, and showing strong robustness to most domain shifts, the exception being a drop in performance on out-of-domain contrasts.
Oriented histogram-based vector field embedding for characterizing 4D CT data sets in radiotherapy
Nov 25, 2024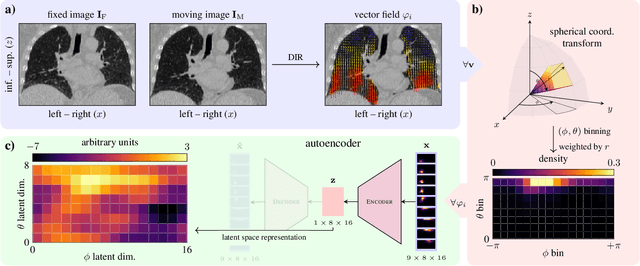
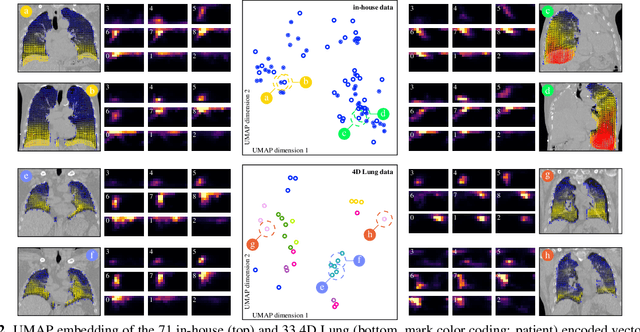
Abstract:In lung radiotherapy, the primary objective is to optimize treatment outcomes by minimizing exposure to healthy tissues while delivering the prescribed dose to the target volume. The challenge lies in accounting for lung tissue motion due to breathing, which impacts precise treatment alignment. To address this, the paper proposes a prospective approach that relies solely on pre-treatment information, such as planning CT scans and derived data like vector fields from deformable image registration. This data is compared to analogous patient data to tailor treatment strategies, i.e., to be able to review treatment parameters and success for similar patients. To allow for such a comparison, an embedding and clustering strategy of prospective patient data is needed. Therefore, the main focus of this study lies on reducing the dimensionality of deformable registration-based vector fields by employing a voxel-wise spherical coordinate transformation and a low-dimensional 2D oriented histogram representation. Afterwards, a fully unsupervised UMAP embedding of the encoded vector fields (i.e., patient-specific motion information) becomes applicable. The functionality of the proposed method is demonstrated with 71 in-house acquired 4D CT data sets and 33 external 4D CT data sets. A comprehensive analysis of the patient clusters is conducted, focusing on the similarity of breathing patterns of clustered patients. The proposed general approach of reducing the dimensionality of registration vector fields by encoding the inherent information into oriented histograms is, however, applicable to other tasks.
Deep learning-based conditional inpainting for restoration of artifact-affected 4D CT images
Mar 12, 2022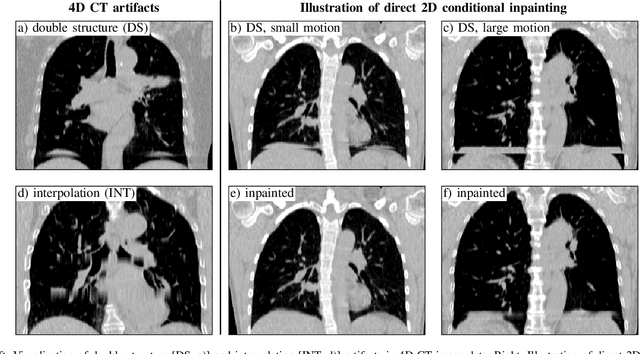
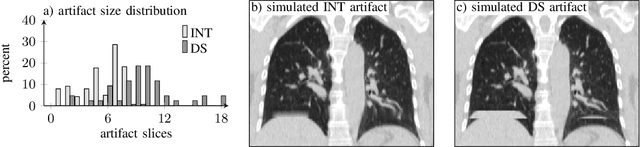
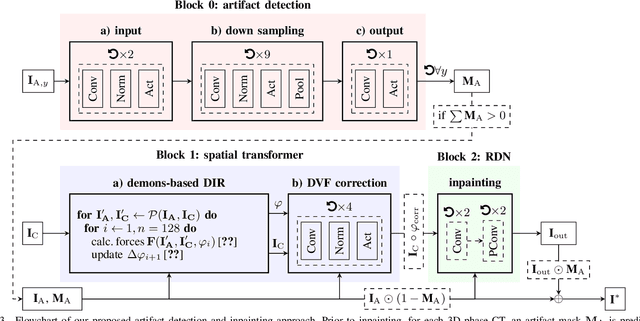
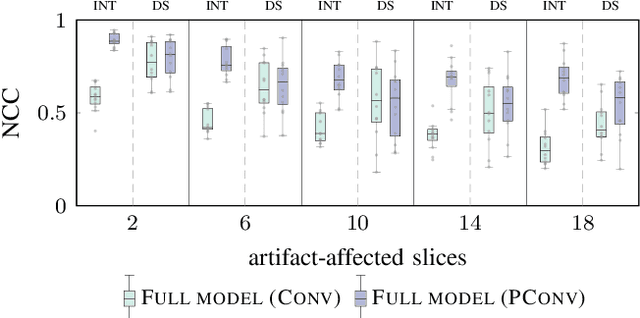
Abstract:4D CT imaging is an essential component of radiotherapy of thoracic/abdominal tumors. 4D CT images are, however, often affected by artifacts that compromise treatment planning quality. In this work, deep learning (DL)-based conditional inpainting is proposed to restore anatomically correct image information of artifact-affected areas. The restoration approach consists of a two-stage process: DL-based detection of common interpolation (INT) and double structure (DS) artifacts, followed by conditional inpainting applied to the artifact areas. In this context, conditional refers to a guidance of the inpainting process by patient-specific image data to ensure anatomically reliable results. Evaluation is based on 65 in-house 4D CT data sets of lung cancer patients (48 with only slight artifacts, 17 with pronounced artifacts) and the publicly available DIRLab 4D CT data (independent external test set). Automated artifact detection revealed a ROC-AUC of 0.99 for INT and 0.97 for DS artifacts (in-house data). The proposed inpainting method decreased the average root mean squared error (RMSE) by 60% (DS) and 42% (INT) for the in-house evaluation data (simulated artifacts for the slight artifact data; original data were considered as ground truth for RMSE computation). For the external DIR-Lab data, the RMSE decreased by 65% and 36%, respectively. Applied to the pronounced artifact data group, on average 68% of the detectable artifacts were removed. The results highlight the potential of DL-based inpainting for the restoration of artifact-affected 4D CT data. Improved performance of conditional inpainting (compared to standard inpainting) illustrates the benefits of exploiting patient-specific prior knowledge.
Multi-scale fully convolutional neural networks for histopathology image segmentation: from nuclear aberrations to the global tissue architecture
Sep 24, 2019
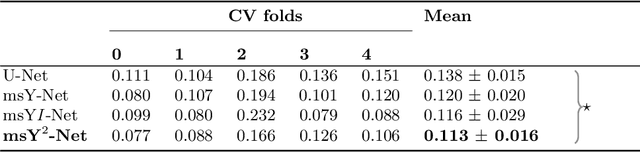
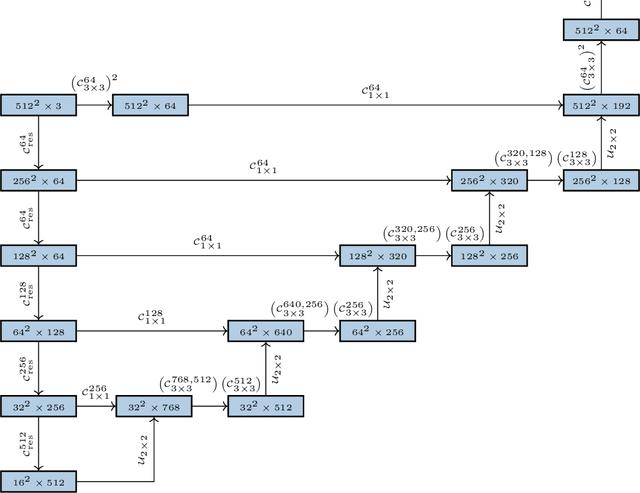
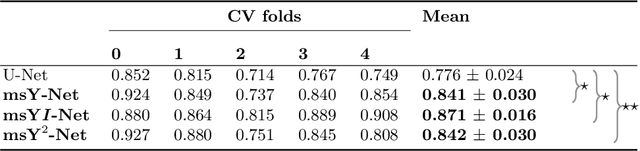
Abstract:Histopathologic diagnosis is dependent on simultaneous information from a broad range of scales, ranging from nuclear aberrations ($\approx \mathcal{O}(0.1 \mu m)$) over cellular structures ($\approx \mathcal{O}(10\mu m)$) to the global tissue architecture ($\gtrapprox \mathcal{O}(1 mm)$). Bearing in mind which information is employed by human pathologists, we introduce and examine different strategies for the integration of multiple and widely separate spatial scales into common U-Net-based architectures. Based on this, we present a family of new, end-to-end trainable, multi-scale multi-encoder fully-convolutional neural networks for human modus operandi-inspired computer vision in histopathology.
Skin Lesion Classification Using CNNs with Patch-Based Attention and Diagnosis-Guided Loss Weighting
May 09, 2019
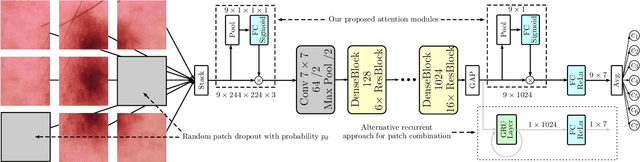
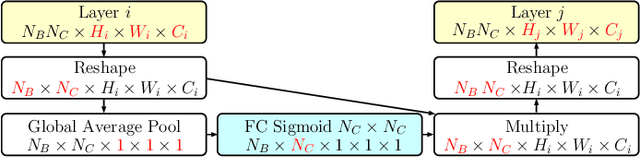
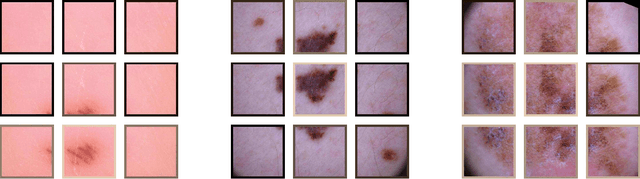
Abstract:Objective: This work addresses two key problems of skin lesion classification. The first problem is the effective use of high-resolution images with pretrained standard architectures for image classification. The second problem is the high class imbalance encountered in real-world multi-class datasets. Methods: To use high-resolution images, we propose a novel patch-based attention architecture that provides global context between small, high-resolution patches. We modify three pretrained architectures and study the performance of patch-based attention. To counter class imbalance problems, we compare oversampling, balanced batch sampling, and class-specific loss weighting. Additionally, we propose a novel diagnosis-guided loss weighting method which takes the method used for ground-truth annotation into account. Results: Our patch-based attention mechanism outperforms previous methods and improves the mean sensitivity by 7%. Class balancing significantly improves the mean sensitivity and we show that our diagnosis-guided loss weighting method improves the mean sensitivity by 3% over normal loss balancing. Conclusion: The novel patch-based attention mechanism can be integrated into pretrained architectures and provides global context between local patches while outperforming other patch-based methods. Hence, pretrained architectures can be readily used with high-resolution images without downsampling. The new diagnosis-guided loss weighting method outperforms other methods and allows for effective training when facing class imbalance. Significance: The proposed methods improve automatic skin lesion classification. They can be extended to other clinical applications where high-resolution image data and class imbalance are relevant.
Skin Lesion Diagnosis using Ensembles, Unscaled Multi-Crop Evaluation and Loss Weighting
Aug 05, 2018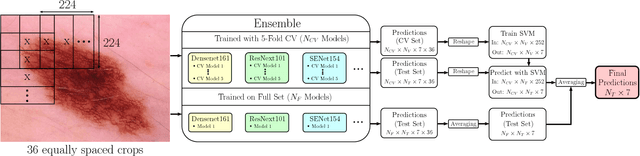
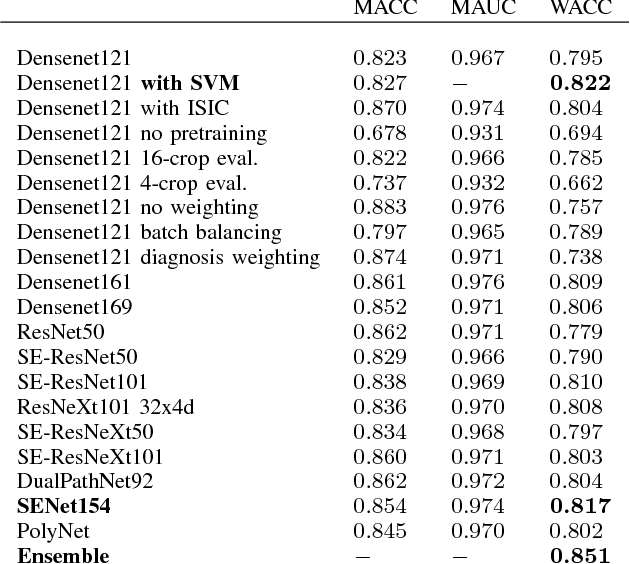
Abstract:In this paper we present the methods of our submission to the ISIC 2018 challenge for skin lesion diagnosis (Task 3). The dataset consists of 10000 images with seven image-level classes to be distinguished by an automated algorithm. We employ an ensemble of convolutional neural networks for this task. In particular, we fine-tune pretrained state-of-the-art deep learning models such as Densenet, SENet and ResNeXt. We identify heavy class imbalance as a key problem for this challenge and consider multiple balancing approaches such as loss weighting and balanced batch sampling. Another important feature of our pipeline is the use of a vast amount of unscaled crops for evaluation. Last, we consider meta learning approaches for the final predictions. Our team placed second at the challenge while being the best approach using only publicly available data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge