Nikola Simidjievski
RO-FIGS: Efficient and Expressive Tree-Based Ensembles for Tabular Data
Apr 09, 2025
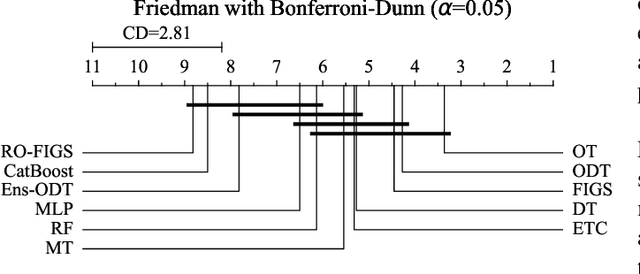
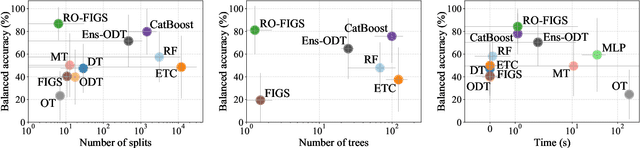
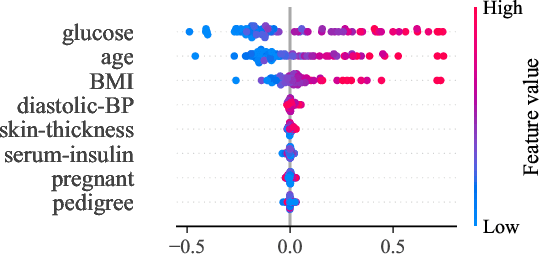
Abstract:Tree-based models are often robust to uninformative features and can accurately capture non-smooth, complex decision boundaries. Consequently, they often outperform neural network-based models on tabular datasets at a significantly lower computational cost. Nevertheless, the capability of traditional tree-based ensembles to express complex relationships efficiently is limited by using a single feature to make splits. To improve the efficiency and expressiveness of tree-based methods, we propose Random Oblique Fast Interpretable Greedy-Tree Sums (RO-FIGS). RO-FIGS builds on Fast Interpretable Greedy-Tree Sums, and extends it by learning trees with oblique or multivariate splits, where each split consists of a linear combination learnt from random subsets of features. This helps uncover interactions between features and improves performance. The proposed method is suitable for tabular datasets with both numerical and categorical features. We evaluate RO-FIGS on 22 real-world tabular datasets, demonstrating superior performance and much smaller models over other tree- and neural network-based methods. Additionally, we analyse their splits to reveal valuable insights into feature interactions, enriching the information learnt from SHAP summary plots, and thereby demonstrating the enhanced interpretability of RO-FIGS models. The proposed method is well-suited for applications, where balance between accuracy and interpretability is essential.
How Well Does Your Tabular Generator Learn the Structure of Tabular Data?
Mar 12, 2025Abstract:Heterogeneous tabular data poses unique challenges in generative modelling due to its fundamentally different underlying data structure compared to homogeneous modalities, such as images and text. Although previous research has sought to adapt the successes of generative modelling in homogeneous modalities to the tabular domain, defining an effective generator for tabular data remains an open problem. One major reason is that the evaluation criteria inherited from other modalities often fail to adequately assess whether tabular generative models effectively capture or utilise the unique structural information encoded in tabular data. In this paper, we carefully examine the limitations of the prevailing evaluation framework and introduce $\textbf{TabStruct}$, a novel evaluation benchmark that positions structural fidelity as a core evaluation dimension. Specifically, TabStruct evaluates the alignment of causal structures in real and synthetic data, providing a direct measure of how effectively tabular generative models learn the structure of tabular data. Through extensive experiments using generators from eight categories on seven datasets with expert-validated causal graphical structures, we show that structural fidelity offers a task-independent, domain-agnostic evaluation dimension. Our findings highlight the importance of tabular data structure and offer practical guidance for developing more effective and robust tabular generative models. Code is available at https://github.com/SilenceX12138/TabStruct.
LLM Embeddings for Deep Learning on Tabular Data
Feb 17, 2025Abstract:Tabular deep-learning methods require embedding numerical and categorical input features into high-dimensional spaces before processing them. Existing methods deal with this heterogeneous nature of tabular data by employing separate type-specific encoding approaches. This limits the cross-table transfer potential and the exploitation of pre-trained knowledge. We propose a novel approach that first transforms tabular data into text, and then leverages pre-trained representations from LLMs to encode this data, resulting in a plug-and-play solution to improv ing deep-learning tabular methods. We demonstrate that our approach improves accuracy over competitive models, such as MLP, ResNet and FT-Transformer, by validating on seven classification datasets.
Measuring Cross-Modal Interactions in Multimodal Models
Dec 20, 2024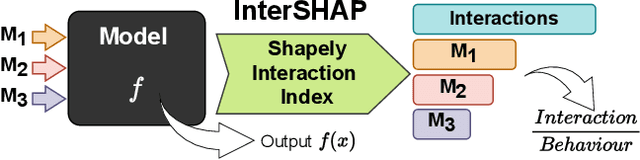
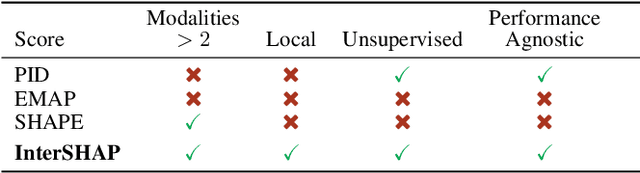
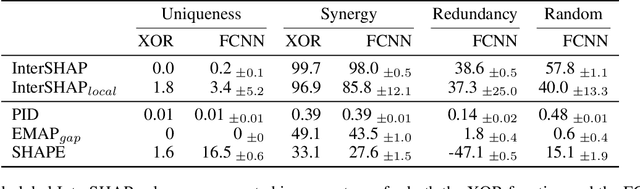
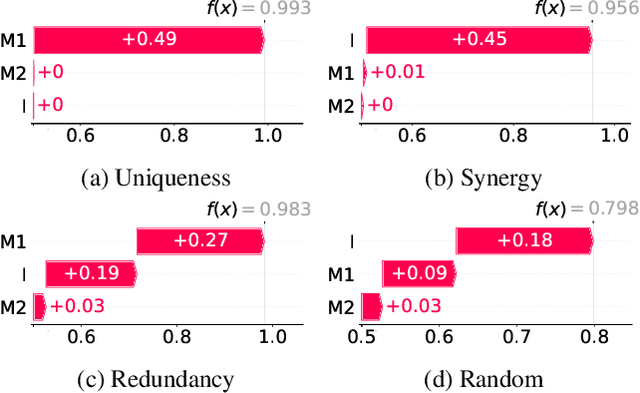
Abstract:Integrating AI in healthcare can greatly improve patient care and system efficiency. However, the lack of explainability in AI systems (XAI) hinders their clinical adoption, especially in multimodal settings that use increasingly complex model architectures. Most existing XAI methods focus on unimodal models, which fail to capture cross-modal interactions crucial for understanding the combined impact of multiple data sources. Existing methods for quantifying cross-modal interactions are limited to two modalities, rely on labelled data, and depend on model performance. This is problematic in healthcare, where XAI must handle multiple data sources and provide individualised explanations. This paper introduces InterSHAP, a cross-modal interaction score that addresses the limitations of existing approaches. InterSHAP uses the Shapley interaction index to precisely separate and quantify the contributions of the individual modalities and their interactions without approximations. By integrating an open-source implementation with the SHAP package, we enhance reproducibility and ease of use. We show that InterSHAP accurately measures the presence of cross-modal interactions, can handle multiple modalities, and provides detailed explanations at a local level for individual samples. Furthermore, we apply InterSHAP to multimodal medical datasets and demonstrate its applicability for individualised explanations.
PATHS: A Hierarchical Transformer for Efficient Whole Slide Image Analysis
Nov 27, 2024Abstract:Computational analysis of whole slide images (WSIs) has seen significant research progress in recent years, with applications ranging across important diagnostic and prognostic tasks such as survival or cancer subtype prediction. Many state-of-the-art models process the entire slide - which may be as large as $150,000 \times 150,000$ pixels - as a bag of many patches, the size of which necessitates computationally cheap feature aggregation methods. However, a large proportion of these patches are uninformative, such as those containing only healthy or adipose tissue, adding significant noise and size to the bag. We propose Pathology Transformer with Hierarchical Selection (PATHS), a novel top-down method for hierarchical weakly supervised representation learning on slide-level tasks in computational pathology. PATHS is inspired by the cross-magnification manner in which a human pathologist examines a slide, recursively filtering patches at each magnification level to a small subset relevant to the diagnosis. Our method overcomes the complications of processing the entire slide, enabling quadratic self-attention and providing a simple interpretable measure of region importance. We apply PATHS to five datasets of The Cancer Genome Atlas (TCGA), and achieve superior performance on slide-level prediction tasks when compared to previous methods, despite processing only a small proportion of the slide.
TabEBM: A Tabular Data Augmentation Method with Distinct Class-Specific Energy-Based Models
Sep 24, 2024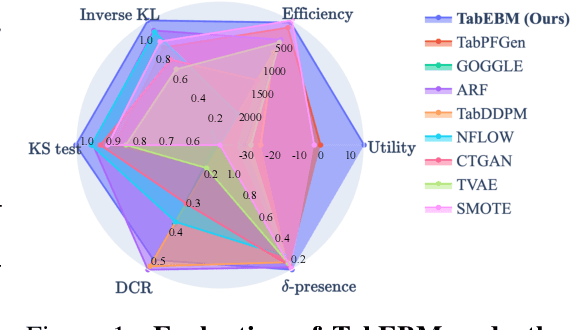
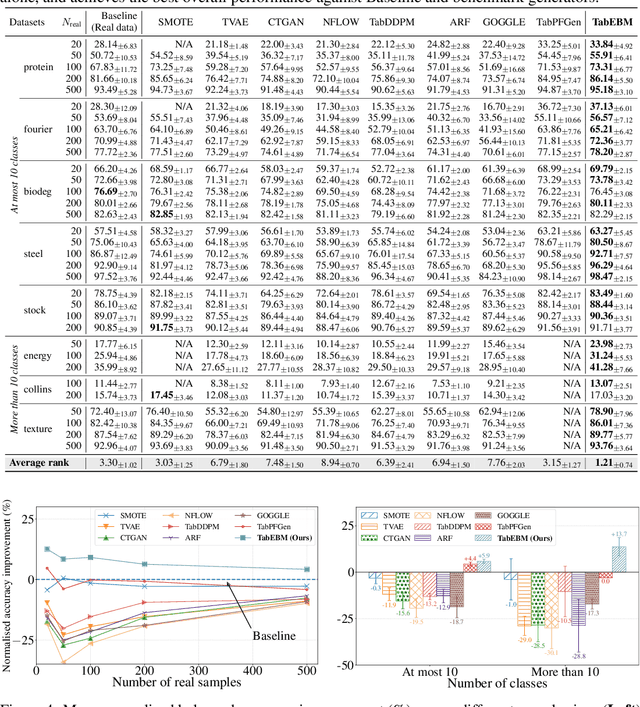
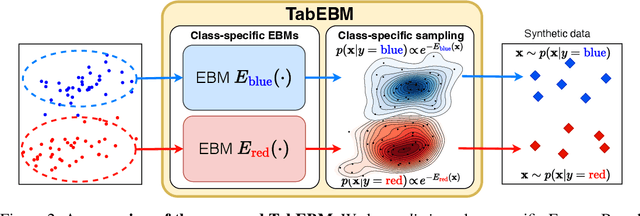
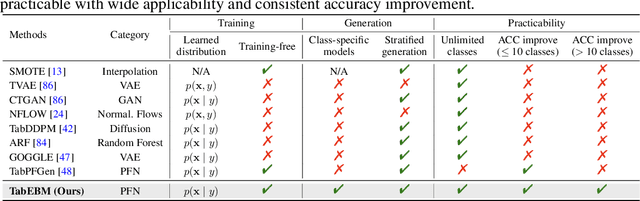
Abstract:Data collection is often difficult in critical fields such as medicine, physics, and chemistry. As a result, classification methods usually perform poorly with these small datasets, leading to weak predictive performance. Increasing the training set with additional synthetic data, similar to data augmentation in images, is commonly believed to improve downstream classification performance. However, current tabular generative methods that learn either the joint distribution $ p(\mathbf{x}, y) $ or the class-conditional distribution $ p(\mathbf{x} \mid y) $ often overfit on small datasets, resulting in poor-quality synthetic data, usually worsening classification performance compared to using real data alone. To solve these challenges, we introduce TabEBM, a novel class-conditional generative method using Energy-Based Models (EBMs). Unlike existing methods that use a shared model to approximate all class-conditional densities, our key innovation is to create distinct EBM generative models for each class, each modelling its class-specific data distribution individually. This approach creates robust energy landscapes, even in ambiguous class distributions. Our experiments show that TabEBM generates synthetic data with higher quality and better statistical fidelity than existing methods. When used for data augmentation, our synthetic data consistently improves the classification performance across diverse datasets of various sizes, especially small ones.
TabMDA: Tabular Manifold Data Augmentation for Any Classifier using Transformers with In-context Subsetting
Jun 03, 2024



Abstract:Tabular data is prevalent in many critical domains, yet it is often challenging to acquire in large quantities. This scarcity usually results in poor performance of machine learning models on such data. Data augmentation, a common strategy for performance improvement in vision and language tasks, typically underperforms for tabular data due to the lack of explicit symmetries in the input space. To overcome this challenge, we introduce TabMDA, a novel method for manifold data augmentation on tabular data. This method utilises a pre-trained in-context model, such as TabPFN, to map the data into a manifold space. TabMDA performs label-invariant transformations by encoding the data multiple times with varied contexts. This process explores the manifold of the underlying in-context models, thereby enlarging the training dataset. TabMDA is a training-free method, making it applicable to any classifier. We evaluate TabMDA on five standard classifiers and observe significant performance improvements across various tabular datasets. Our results demonstrate that TabMDA provides an effective way to leverage information from pre-trained in-context models to enhance the performance of downstream classifiers.
MM-Lego: Modular Biomedical Multimodal Models with Minimal Fine-Tuning
May 30, 2024



Abstract:Learning holistic computational representations in physical, chemical or biological systems requires the ability to process information from different distributions and modalities within the same model. Thus, the demand for multimodal machine learning models has sharply risen for modalities that go beyond vision and language, such as sequences, graphs, time series, or tabular data. While there are many available multimodal fusion and alignment approaches, most of them require end-to-end training, scale quadratically with the number of modalities, cannot handle cases of high modality imbalance in the training set, or are highly topology-specific, making them too restrictive for many biomedical learning tasks. This paper presents Multimodal Lego (MM-Lego), a modular and general-purpose fusion and model merging framework to turn any set of encoders into a competitive multimodal model with no or minimal fine-tuning. We achieve this by introducing a wrapper for unimodal encoders that enforces lightweight dimensionality assumptions between modalities and harmonises their representations by learning features in the frequency domain to enable model merging with little signal interference. We show that MM-Lego 1) can be used as a model merging method which achieves competitive performance with end-to-end fusion models without any fine-tuning, 2) can operate on any unimodal encoder, and 3) is a model fusion method that, with minimal fine-tuning, achieves state-of-the-art results on six benchmarked multimodal biomedical tasks.
Everybody Needs a Little HELP: Explaining Graphs via Hierarchical Concepts
Dec 02, 2023


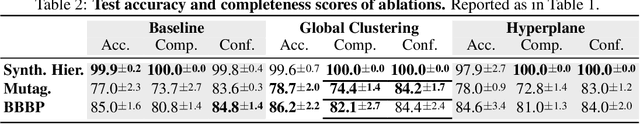
Abstract:Graph neural networks (GNNs) have led to major breakthroughs in a variety of domains such as drug discovery, social network analysis, and travel time estimation. However, they lack interpretability which hinders human trust and thereby deployment to settings with high-stakes decisions. A line of interpretable methods approach this by discovering a small set of relevant concepts as subgraphs in the last GNN layer that together explain the prediction. This can yield oversimplified explanations, failing to explain the interaction between GNN layers. To address this oversight, we provide HELP (Hierarchical Explainable Latent Pooling), a novel, inherently interpretable graph pooling approach that reveals how concepts from different GNN layers compose to new ones in later steps. HELP is more than 1-WL expressive and is the first non-spectral, end-to-end-learnable, hierarchical graph pooling method that can learn to pool a variable number of arbitrary connected components. We empirically demonstrate that it performs on-par with standard GCNs and popular pooling methods in terms of accuracy while yielding explanations that are aligned with expert knowledge in the domains of chemistry and social networks. In addition to a qualitative analysis, we employ concept completeness scores as well as concept conformity, a novel metric to measure the noise in discovered concepts, quantitatively verifying that the discovered concepts are significantly easier to fully understand than those from previous work. Our work represents a first step towards an understanding of graph neural networks that goes beyond a set of concepts from the final layer and instead explains the complex interplay of concepts on different levels.
HEALNet -- Hybrid Multi-Modal Fusion for Heterogeneous Biomedical Data
Nov 20, 2023



Abstract:Technological advances in medical data collection such as high-resolution histopathology and high-throughput genomic sequencing have contributed to the rising requirement for multi-modal biomedical modelling, specifically for image, tabular, and graph data. Most multi-modal deep learning approaches use modality-specific architectures that are trained separately and cannot capture the crucial cross-modal information that motivates the integration of different data sources. This paper presents the Hybrid Early-fusion Attention Learning Network (HEALNet): a flexible multi-modal fusion architecture, which a) preserves modality-specific structural information, b) captures the cross-modal interactions and structural information in a shared latent space, c) can effectively handle missing modalities during training and inference, and d) enables intuitive model inspection by learning on the raw data input instead of opaque embeddings. We conduct multi-modal survival analysis on Whole Slide Images and Multi-omic data on four cancer cohorts of The Cancer Genome Atlas (TCGA). HEALNet achieves state-of-the-art performance, substantially improving over both uni-modal and recent multi-modal baselines, whilst being robust in scenarios with missing modalities.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge