Andrei Margeloiu
LLM Embeddings for Deep Learning on Tabular Data
Feb 17, 2025Abstract:Tabular deep-learning methods require embedding numerical and categorical input features into high-dimensional spaces before processing them. Existing methods deal with this heterogeneous nature of tabular data by employing separate type-specific encoding approaches. This limits the cross-table transfer potential and the exploitation of pre-trained knowledge. We propose a novel approach that first transforms tabular data into text, and then leverages pre-trained representations from LLMs to encode this data, resulting in a plug-and-play solution to improv ing deep-learning tabular methods. We demonstrate that our approach improves accuracy over competitive models, such as MLP, ResNet and FT-Transformer, by validating on seven classification datasets.
TabEBM: A Tabular Data Augmentation Method with Distinct Class-Specific Energy-Based Models
Sep 24, 2024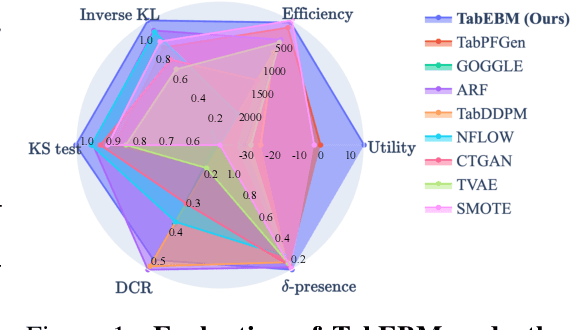
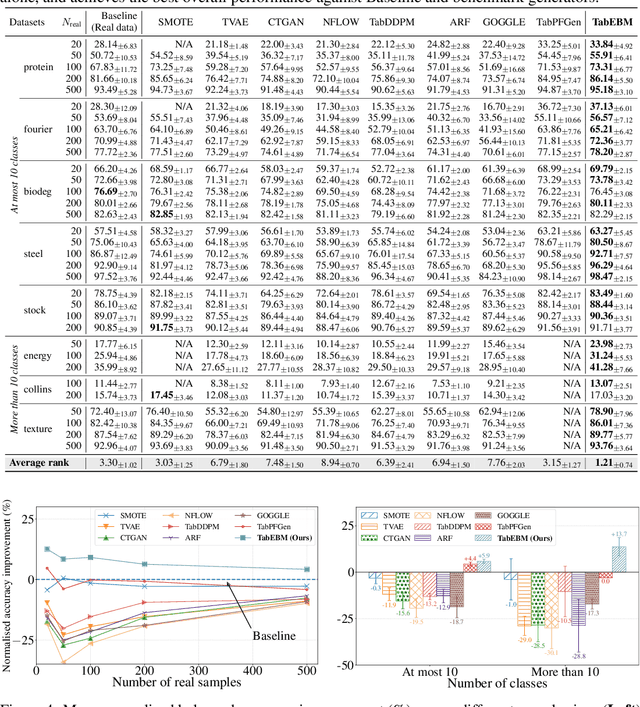
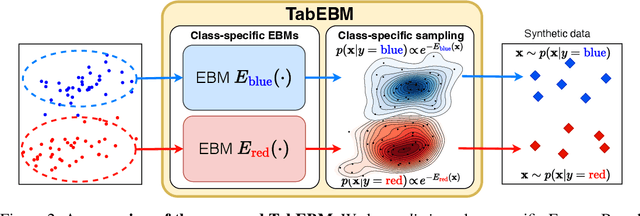
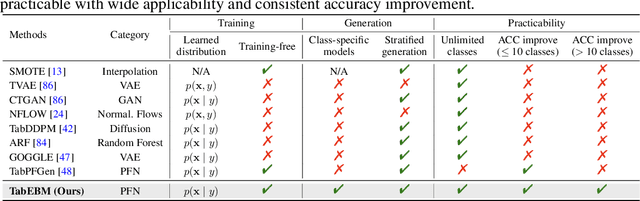
Abstract:Data collection is often difficult in critical fields such as medicine, physics, and chemistry. As a result, classification methods usually perform poorly with these small datasets, leading to weak predictive performance. Increasing the training set with additional synthetic data, similar to data augmentation in images, is commonly believed to improve downstream classification performance. However, current tabular generative methods that learn either the joint distribution $ p(\mathbf{x}, y) $ or the class-conditional distribution $ p(\mathbf{x} \mid y) $ often overfit on small datasets, resulting in poor-quality synthetic data, usually worsening classification performance compared to using real data alone. To solve these challenges, we introduce TabEBM, a novel class-conditional generative method using Energy-Based Models (EBMs). Unlike existing methods that use a shared model to approximate all class-conditional densities, our key innovation is to create distinct EBM generative models for each class, each modelling its class-specific data distribution individually. This approach creates robust energy landscapes, even in ambiguous class distributions. Our experiments show that TabEBM generates synthetic data with higher quality and better statistical fidelity than existing methods. When used for data augmentation, our synthetic data consistently improves the classification performance across diverse datasets of various sizes, especially small ones.
TabMDA: Tabular Manifold Data Augmentation for Any Classifier using Transformers with In-context Subsetting
Jun 03, 2024



Abstract:Tabular data is prevalent in many critical domains, yet it is often challenging to acquire in large quantities. This scarcity usually results in poor performance of machine learning models on such data. Data augmentation, a common strategy for performance improvement in vision and language tasks, typically underperforms for tabular data due to the lack of explicit symmetries in the input space. To overcome this challenge, we introduce TabMDA, a novel method for manifold data augmentation on tabular data. This method utilises a pre-trained in-context model, such as TabPFN, to map the data into a manifold space. TabMDA performs label-invariant transformations by encoding the data multiple times with varied contexts. This process explores the manifold of the underlying in-context models, thereby enlarging the training dataset. TabMDA is a training-free method, making it applicable to any classifier. We evaluate TabMDA on five standard classifiers and observe significant performance improvements across various tabular datasets. Our results demonstrate that TabMDA provides an effective way to leverage information from pre-trained in-context models to enhance the performance of downstream classifiers.
Enhancing Representation Learning on High-Dimensional, Small-Size Tabular Data: A Divide and Conquer Method with Ensembled VAEs
Jun 27, 2023



Abstract:Variational Autoencoders and their many variants have displayed impressive ability to perform dimensionality reduction, often achieving state-of-the-art performance. Many current methods however, struggle to learn good representations in High Dimensional, Low Sample Size (HDLSS) tasks, which is an inherently challenging setting. We address this challenge by using an ensemble of lightweight VAEs to learn posteriors over subsets of the feature-space, which get aggregated into a joint posterior in a novel divide-and-conquer approach. Specifically, we present an alternative factorisation of the joint posterior that induces a form of implicit data augmentation that yields greater sample efficiency. Through a series of experiments on eight real-world datasets, we show that our method learns better latent representations in HDLSS settings, which leads to higher accuracy in a downstream classification task. Furthermore, we verify that our approach has a positive effect on disentanglement and achieves a lower estimated Total Correlation on learnt representations. Finally, we show that our approach is robust to partial features at inference, exhibiting little performance degradation even with most features missing.
ProtoGate: Prototype-based Neural Networks with Local Feature Selection for Tabular Biomedical Data
Jun 21, 2023


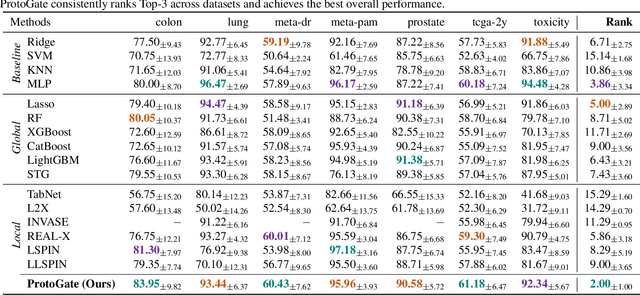
Abstract:Tabular biomedical data poses challenges in machine learning because it is often high-dimensional and typically low-sample-size. Previous research has attempted to address these challenges via feature selection approaches, which can lead to unstable performance on real-world data. This suggests that current methods lack appropriate inductive biases that capture patterns common to different samples. In this paper, we propose ProtoGate, a prototype-based neural model that introduces an inductive bias by attending to both homogeneity and heterogeneity across samples. ProtoGate selects features in a global-to-local manner and leverages them to produce explainable predictions via an interpretable prototype-based model. We conduct comprehensive experiments to evaluate the performance of ProtoGate on synthetic and real-world datasets. Our results show that exploiting the homogeneous and heterogeneous patterns in the data can improve prediction accuracy while prototypes imbue interpretability.
Weight Predictor Network with Feature Selection for Small Sample Tabular Biomedical Data
Nov 28, 2022Abstract:Tabular biomedical data is often high-dimensional but with a very small number of samples. Although recent work showed that well-regularised simple neural networks could outperform more sophisticated architectures on tabular data, they are still prone to overfitting on tiny datasets with many potentially irrelevant features. To combat these issues, we propose Weight Predictor Network with Feature Selection (WPFS) for learning neural networks from high-dimensional and small sample data by reducing the number of learnable parameters and simultaneously performing feature selection. In addition to the classification network, WPFS uses two small auxiliary networks that together output the weights of the first layer of the classification model. We evaluate on nine real-world biomedical datasets and demonstrate that WPFS outperforms other standard as well as more recent methods typically applied to tabular data. Furthermore, we investigate the proposed feature selection mechanism and show that it improves performance while providing useful insights into the learning task.
Graph-Conditioned MLP for High-Dimensional Tabular Biomedical Data
Nov 11, 2022



Abstract:Genome-wide studies leveraging recent high-throughput sequencing technologies collect high-dimensional data. However, they usually include small cohorts of patients, and the resulting tabular datasets suffer from the "curse of dimensionality". Training neural networks on such datasets is typically unstable, and the models overfit. One problem is that modern weight initialisation strategies make simplistic assumptions unsuitable for small-size datasets. We propose Graph-Conditioned MLP, a novel method to introduce priors on the parameters of an MLP. Instead of randomly initialising the first layer, we condition it directly on the training data. More specifically, we create a graph for each feature in the dataset (e.g., a gene), where each node represents a sample from the same dataset (e.g., a patient). We then use Graph Neural Networks (GNNs) to learn embeddings from these graphs and use the embeddings to initialise the MLP's parameters. Our approach opens the prospect of introducing additional biological knowledge when constructing the graphs. We present early results on 7 classification tasks from gene expression data and show that GC-MLP outperforms an MLP.
Do Concept Bottleneck Models Learn as Intended?
May 10, 2021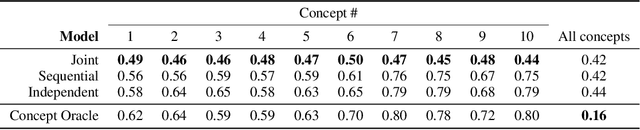
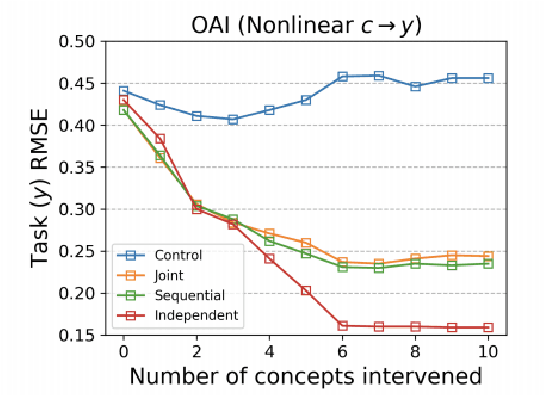
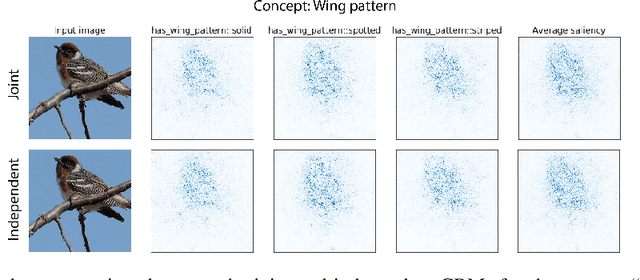
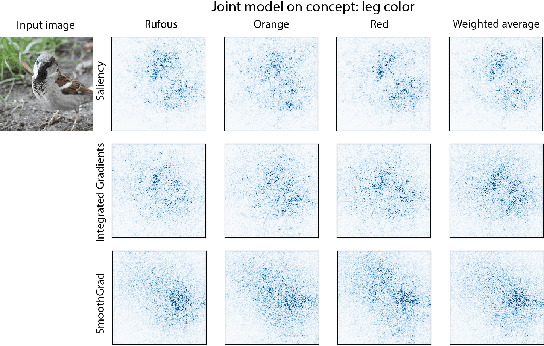
Abstract:Concept bottleneck models map from raw inputs to concepts, and then from concepts to targets. Such models aim to incorporate pre-specified, high-level concepts into the learning procedure, and have been motivated to meet three desiderata: interpretability, predictability, and intervenability. However, we find that concept bottleneck models struggle to meet these goals. Using post hoc interpretability methods, we demonstrate that concepts do not correspond to anything semantically meaningful in input space, thus calling into question the usefulness of concept bottleneck models in their current form.
Improving Interpretability in Medical Imaging Diagnosis using Adversarial Training
Dec 02, 2020



Abstract:We investigate the influence of adversarial training on the interpretability of convolutional neural networks (CNNs), specifically applied to diagnosing skin cancer. We show that gradient-based saliency maps of adversarially trained CNNs are significantly sharper and more visually coherent than those of standardly trained CNNs. Furthermore, we show that adversarially trained networks highlight regions with significant color variation within the lesion, a common characteristic of melanoma. We find that fine-tuning a robust network with a small learning rate further improves saliency maps' sharpness. Lastly, we provide preliminary work suggesting that robustifying the first layers to extract robust low-level features leads to visually coherent explanations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge