Tong Ding
Parameter estimation of range-migrating targets using OTFS signals from LEO satellites
Jul 03, 2025Abstract:This study investigates a communication-centric integrated sensing and communication (ISAC) system that utilizes orthogonal time frequency space (OTFS) modulated signals emitted by low Earth orbit (LEO) satellites to estimate the parameters of space targets experiencing range migration, henceforth referred to as high-speed targets. Leveraging the specific signal processing performed by OTFS transceivers, we derive a novel input-output model for the echo generated by a high-speed target in scenarios where ideal and rectangular shaping filters are employed. Our findings reveal that the target response exhibits a sparse structure in the delay-Doppler domain, dependent solely upon the initial range and range-rate; notably, range migration causes a spread in the target response, marking a significant departure from previous studies. Utilizing this signal structure, we propose an approximate implementation of the maximum likelihood estimator for the target's initial range, range-rate, and amplitude. The estimation process involves obtaining coarse information on the target response using a block orthogonal matching pursuit algorithm, followed by a refinement step using a bank of matched filters focused on a smaller range and range-rate region. Finally, numerical examples are provided to evaluate the estimation performance.
Evidence-based diagnostic reasoning with multi-agent copilot for human pathology
Jun 26, 2025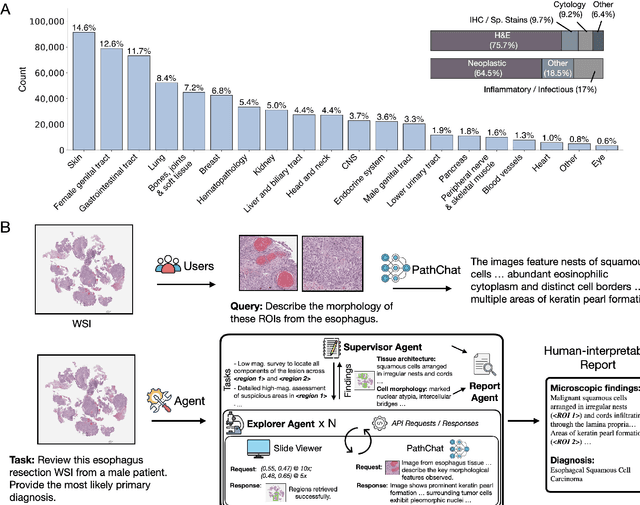
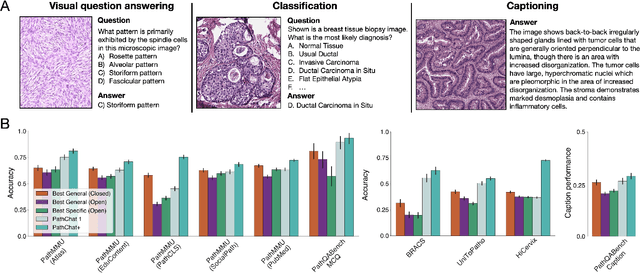
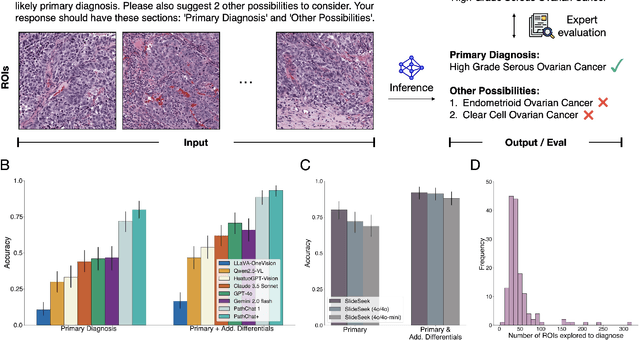
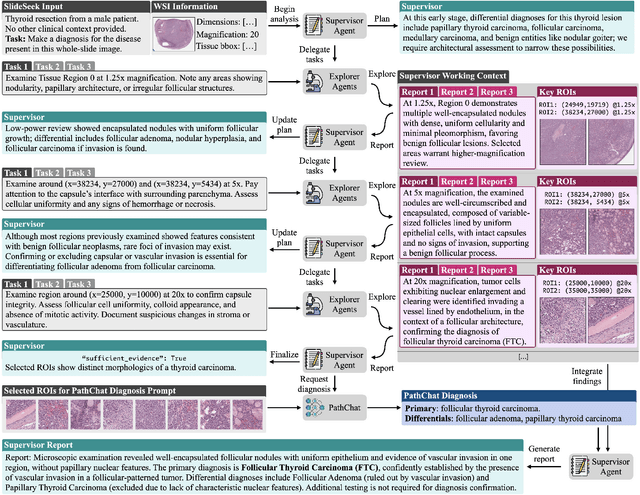
Abstract:Pathology is experiencing rapid digital transformation driven by whole-slide imaging and artificial intelligence (AI). While deep learning-based computational pathology has achieved notable success, traditional models primarily focus on image analysis without integrating natural language instruction or rich, text-based context. Current multimodal large language models (MLLMs) in computational pathology face limitations, including insufficient training data, inadequate support and evaluation for multi-image understanding, and a lack of autonomous, diagnostic reasoning capabilities. To address these limitations, we introduce PathChat+, a new MLLM specifically designed for human pathology, trained on over 1 million diverse, pathology-specific instruction samples and nearly 5.5 million question answer turns. Extensive evaluations across diverse pathology benchmarks demonstrated that PathChat+ substantially outperforms the prior PathChat copilot, as well as both state-of-the-art (SOTA) general-purpose and other pathology-specific models. Furthermore, we present SlideSeek, a reasoning-enabled multi-agent AI system leveraging PathChat+ to autonomously evaluate gigapixel whole-slide images (WSIs) through iterative, hierarchical diagnostic reasoning, reaching high accuracy on DDxBench, a challenging open-ended differential diagnosis benchmark, while also capable of generating visually grounded, humanly-interpretable summary reports.
Do Multiple Instance Learning Models Transfer?
Jun 11, 2025Abstract:Multiple Instance Learning (MIL) is a cornerstone approach in computational pathology (CPath) for generating clinically meaningful slide-level embeddings from gigapixel tissue images. However, MIL often struggles with small, weakly supervised clinical datasets. In contrast to fields such as NLP and conventional computer vision, where transfer learning is widely used to address data scarcity, the transferability of MIL models remains poorly understood. In this study, we systematically evaluate the transfer learning capabilities of pretrained MIL models by assessing 11 models across 21 pretraining tasks for morphological and molecular subtype prediction. Our results show that pretrained MIL models, even when trained on different organs than the target task, consistently outperform models trained from scratch. Moreover, pretraining on pancancer datasets enables strong generalization across organs and tasks, outperforming slide foundation models while using substantially less pretraining data. These findings highlight the robust adaptability of MIL models and demonstrate the benefits of leveraging transfer learning to boost performance in CPath. Lastly, we provide a resource which standardizes the implementation of MIL models and collection of pretrained model weights on popular CPath tasks, available at https://github.com/mahmoodlab/MIL-Lab
Accelerating Data Processing and Benchmarking of AI Models for Pathology
Feb 10, 2025Abstract:Advances in foundation modeling have reshaped computational pathology. However, the increasing number of available models and lack of standardized benchmarks make it increasingly complex to assess their strengths, limitations, and potential for further development. To address these challenges, we introduce a new suite of software tools for whole-slide image processing, foundation model benchmarking, and curated publicly available tasks. We anticipate that these resources will promote transparency, reproducibility, and continued progress in the field.
Molecular-driven Foundation Model for Oncologic Pathology
Jan 28, 2025Abstract:Foundation models are reshaping computational pathology by enabling transfer learning, where models pre-trained on vast datasets can be adapted for downstream diagnostic, prognostic, and therapeutic response tasks. Despite these advances, foundation models are still limited in their ability to encode the entire gigapixel whole-slide images without additional training and often lack complementary multimodal data. Here, we introduce Threads, a slide-level foundation model capable of generating universal representations of whole-slide images of any size. Threads was pre-trained using a multimodal learning approach on a diverse cohort of 47,171 hematoxylin and eosin (H&E)-stained tissue sections, paired with corresponding genomic and transcriptomic profiles - the largest such paired dataset to be used for foundation model development to date. This unique training paradigm enables Threads to capture the tissue's underlying molecular composition, yielding powerful representations applicable to a wide array of downstream tasks. In extensive benchmarking across 54 oncology tasks, including clinical subtyping, grading, mutation prediction, immunohistochemistry status determination, treatment response prediction, and survival prediction, Threads outperformed all baselines while demonstrating remarkable generalizability and label efficiency. It is particularly well suited for predicting rare events, further emphasizing its clinical utility. We intend to make the model publicly available for the broader community.
Multimodal Whole Slide Foundation Model for Pathology
Nov 29, 2024



Abstract:The field of computational pathology has been transformed with recent advances in foundation models that encode histopathology region-of-interests (ROIs) into versatile and transferable feature representations via self-supervised learning (SSL). However, translating these advancements to address complex clinical challenges at the patient and slide level remains constrained by limited clinical data in disease-specific cohorts, especially for rare clinical conditions. We propose TITAN, a multimodal whole slide foundation model pretrained using 335,645 WSIs via visual self-supervised learning and vision-language alignment with corresponding pathology reports and 423,122 synthetic captions generated from a multimodal generative AI copilot for pathology. Without any finetuning or requiring clinical labels, TITAN can extract general-purpose slide representations and generate pathology reports that generalize to resource-limited clinical scenarios such as rare disease retrieval and cancer prognosis. We evaluate TITAN on diverse clinical tasks and find that TITAN outperforms both ROI and slide foundation models across machine learning settings such as linear probing, few-shot and zero-shot classification, rare cancer retrieval and cross-modal retrieval, and pathology report generation.
Tree of Attributes Prompt Learning for Vision-Language Models
Oct 15, 2024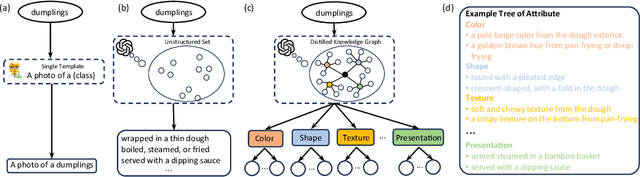
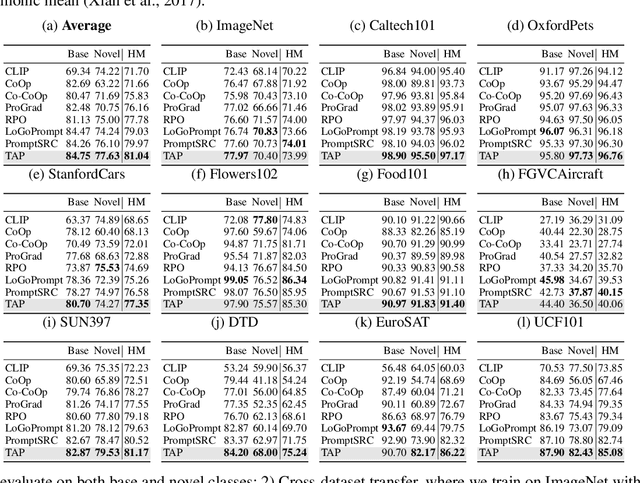
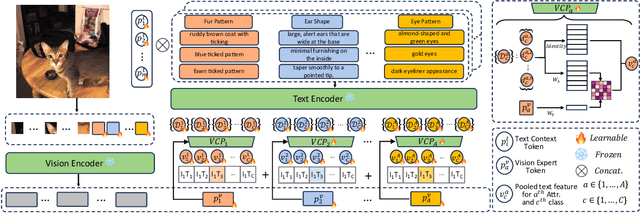
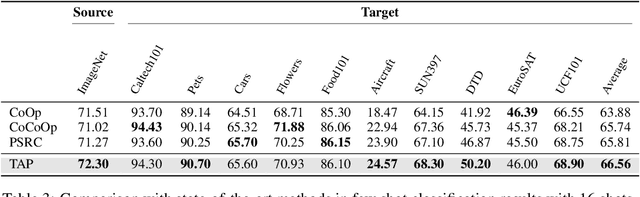
Abstract:Prompt learning has proven effective in adapting vision language models for downstream tasks. However, existing methods usually append learnable prompt tokens solely with the category names to obtain textual features, which fails to fully leverage the rich context indicated in the category name. To address this issue, we propose the Tree of Attributes Prompt learning (TAP), which first instructs LLMs to generate a tree of attributes with a "concept - attribute - description" structure for each category, and then learn the hierarchy with vision and text prompt tokens. Unlike existing methods that merely augment category names with a set of unstructured descriptions, our approach essentially distills structured knowledge graphs associated with class names from LLMs. Furthermore, our approach introduces text and vision prompts designed to explicitly learn the corresponding visual attributes, effectively serving as domain experts. Additionally, the general and diverse descriptions generated based on the class names may be wrong or absent in the specific given images. To address this misalignment, we further introduce a vision-conditional pooling module to extract instance-specific text features. Extensive experimental results demonstrate that our approach outperforms state-of-the-art methods on the zero-shot base-to-novel generalization, cross-dataset transfer, as well as few-shot classification across 11 diverse datasets.
Morphological Prototyping for Unsupervised Slide Representation Learning in Computational Pathology
May 19, 2024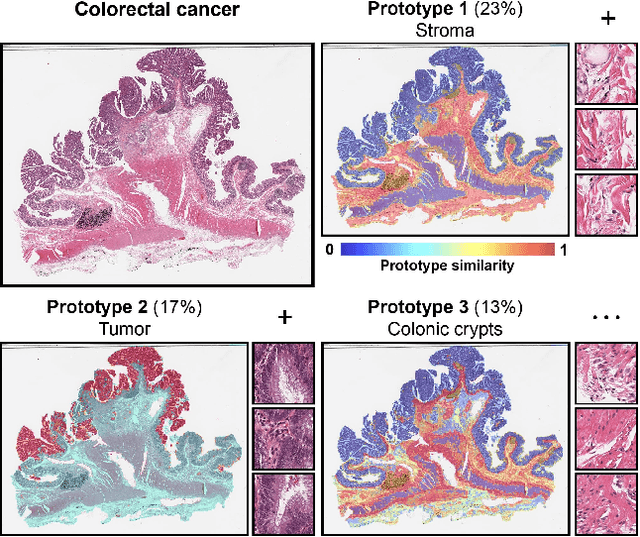
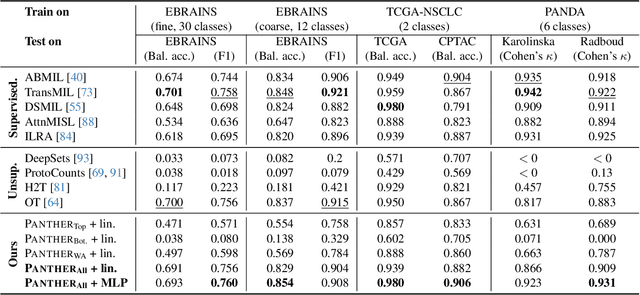
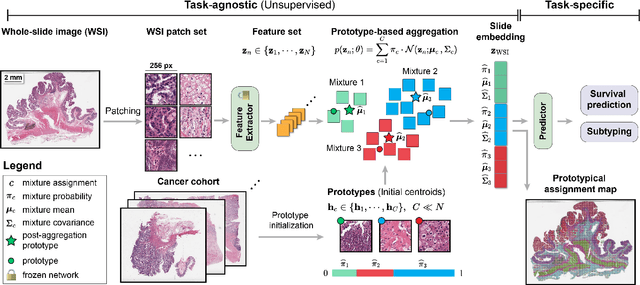
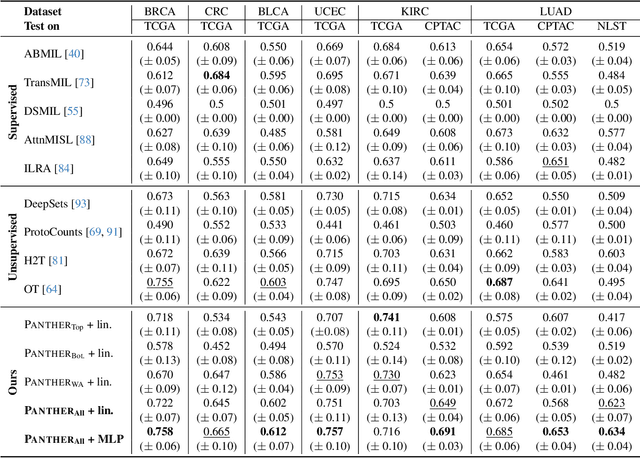
Abstract:Representation learning of pathology whole-slide images (WSIs) has been has primarily relied on weak supervision with Multiple Instance Learning (MIL). However, the slide representations resulting from this approach are highly tailored to specific clinical tasks, which limits their expressivity and generalization, particularly in scenarios with limited data. Instead, we hypothesize that morphological redundancy in tissue can be leveraged to build a task-agnostic slide representation in an unsupervised fashion. To this end, we introduce PANTHER, a prototype-based approach rooted in the Gaussian mixture model that summarizes the set of WSI patches into a much smaller set of morphological prototypes. Specifically, each patch is assumed to have been generated from a mixture distribution, where each mixture component represents a morphological exemplar. Utilizing the estimated mixture parameters, we then construct a compact slide representation that can be readily used for a wide range of downstream tasks. By performing an extensive evaluation of PANTHER on subtyping and survival tasks using 13 datasets, we show that 1) PANTHER outperforms or is on par with supervised MIL baselines and 2) the analysis of morphological prototypes brings new qualitative and quantitative insights into model interpretability.
A Structure-Guided Gauss-Newton Method for Shallow ReLU Neural Network
Apr 07, 2024Abstract:In this paper, we propose a structure-guided Gauss-Newton (SgGN) method for solving least squares problems using a shallow ReLU neural network. The method effectively takes advantage of both the least squares structure and the neural network structure of the objective function. By categorizing the weights and biases of the hidden and output layers of the network as nonlinear and linear parameters, respectively, the method iterates back and forth between the nonlinear and linear parameters. The nonlinear parameters are updated by a damped Gauss-Newton method and the linear ones are updated by a linear solver. Moreover, at the Gauss-Newton step, a special form of the Gauss-Newton matrix is derived for the shallow ReLU neural network and is used for efficient iterations. It is shown that the corresponding mass and Gauss-Newton matrices in the respective linear and nonlinear steps are symmetric and positive definite under reasonable assumptions. Thus, the SgGN method naturally produces an effective search direction without the need of additional techniques like shifting in the Levenberg-Marquardt method to achieve invertibility of the Gauss-Newton matrix. The convergence and accuracy of the method are demonstrated numerically for several challenging function approximation problems, especially those with discontinuities or sharp transition layers that pose significant challenges for commonly used training algorithms in machine learning.
A Foundational Multimodal Vision Language AI Assistant for Human Pathology
Dec 13, 2023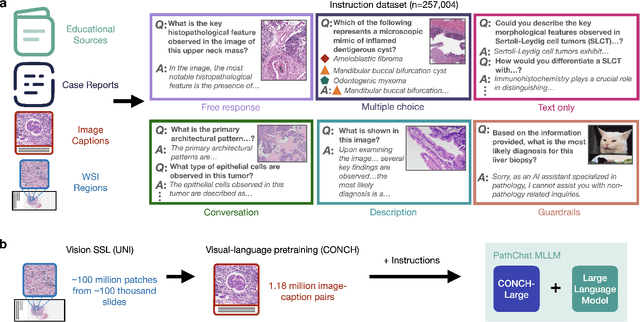
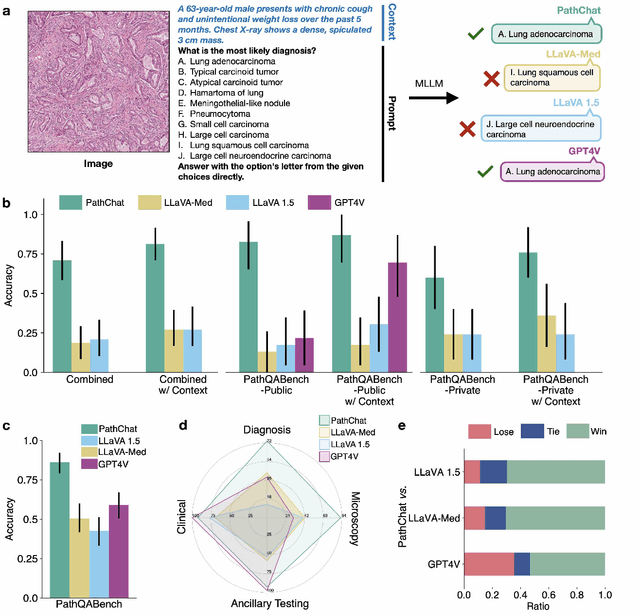
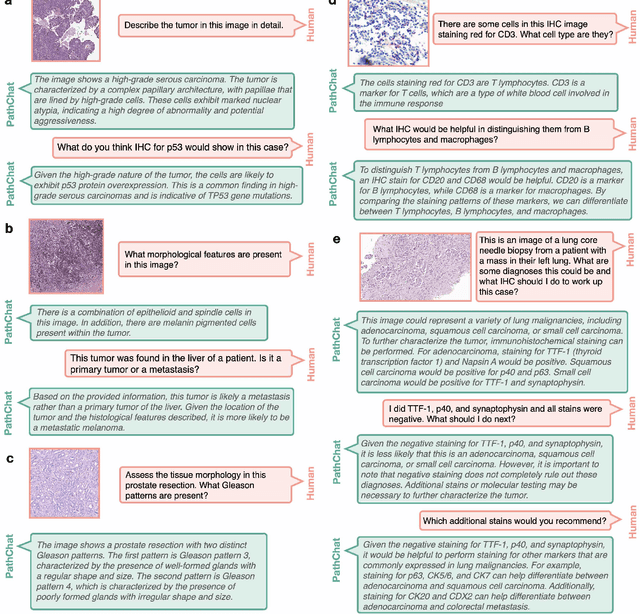
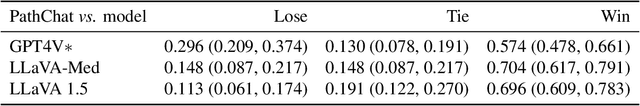
Abstract:The field of computational pathology has witnessed remarkable progress in the development of both task-specific predictive models and task-agnostic self-supervised vision encoders. However, despite the explosive growth of generative artificial intelligence (AI), there has been limited study on building general purpose, multimodal AI assistants tailored to pathology. Here we present PathChat, a vision-language generalist AI assistant for human pathology using an in-house developed foundational vision encoder pretrained on 100 million histology images from over 100,000 patient cases and 1.18 million pathology image-caption pairs. The vision encoder is then combined with a pretrained large language model and the whole system is finetuned on over 250,000 diverse disease agnostic visual language instructions. We compare PathChat against several multimodal vision language AI assistants as well as GPT4V, which powers the commercially available multimodal general purpose AI assistant ChatGPT-4. When relevant clinical context is provided with the histology image, PathChat achieved a diagnostic accuracy of 87% on multiple-choice questions based on publicly available cases of diverse tissue origins and disease models. Additionally, using open-ended questions and human expert evaluation, we found that overall PathChat produced more accurate and pathologist-preferable responses to diverse queries related to pathology. As an interactive and general vision language AI assistant that can flexibly handle both visual and natural language inputs, PathChat can potentially find impactful applications in pathology education, research, and human-in-the-loop clinical decision making.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge