Georg Gerber
Molecular-driven Foundation Model for Oncologic Pathology
Jan 28, 2025Abstract:Foundation models are reshaping computational pathology by enabling transfer learning, where models pre-trained on vast datasets can be adapted for downstream diagnostic, prognostic, and therapeutic response tasks. Despite these advances, foundation models are still limited in their ability to encode the entire gigapixel whole-slide images without additional training and often lack complementary multimodal data. Here, we introduce Threads, a slide-level foundation model capable of generating universal representations of whole-slide images of any size. Threads was pre-trained using a multimodal learning approach on a diverse cohort of 47,171 hematoxylin and eosin (H&E)-stained tissue sections, paired with corresponding genomic and transcriptomic profiles - the largest such paired dataset to be used for foundation model development to date. This unique training paradigm enables Threads to capture the tissue's underlying molecular composition, yielding powerful representations applicable to a wide array of downstream tasks. In extensive benchmarking across 54 oncology tasks, including clinical subtyping, grading, mutation prediction, immunohistochemistry status determination, treatment response prediction, and survival prediction, Threads outperformed all baselines while demonstrating remarkable generalizability and label efficiency. It is particularly well suited for predicting rare events, further emphasizing its clinical utility. We intend to make the model publicly available for the broader community.
Multimodal Whole Slide Foundation Model for Pathology
Nov 29, 2024



Abstract:The field of computational pathology has been transformed with recent advances in foundation models that encode histopathology region-of-interests (ROIs) into versatile and transferable feature representations via self-supervised learning (SSL). However, translating these advancements to address complex clinical challenges at the patient and slide level remains constrained by limited clinical data in disease-specific cohorts, especially for rare clinical conditions. We propose TITAN, a multimodal whole slide foundation model pretrained using 335,645 WSIs via visual self-supervised learning and vision-language alignment with corresponding pathology reports and 423,122 synthetic captions generated from a multimodal generative AI copilot for pathology. Without any finetuning or requiring clinical labels, TITAN can extract general-purpose slide representations and generate pathology reports that generalize to resource-limited clinical scenarios such as rare disease retrieval and cancer prognosis. We evaluate TITAN on diverse clinical tasks and find that TITAN outperforms both ROI and slide foundation models across machine learning settings such as linear probing, few-shot and zero-shot classification, rare cancer retrieval and cross-modal retrieval, and pathology report generation.
A Foundational Multimodal Vision Language AI Assistant for Human Pathology
Dec 13, 2023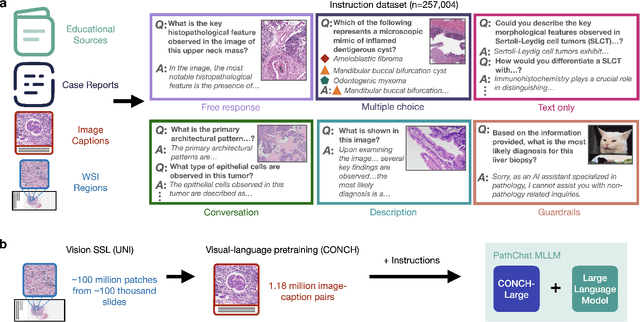
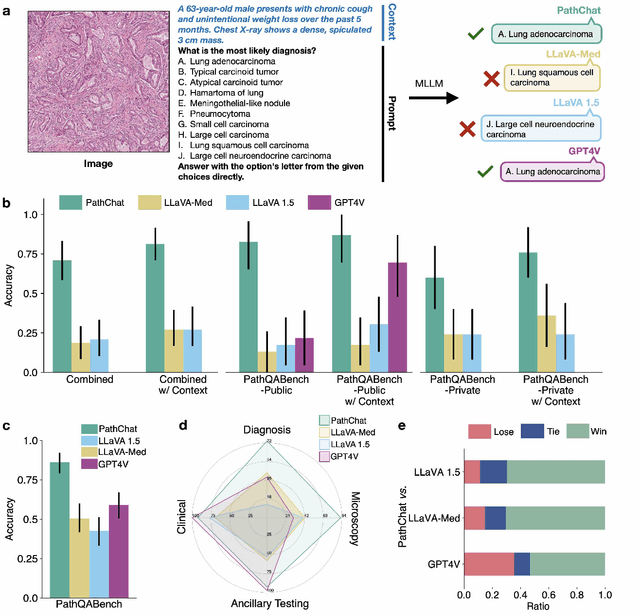
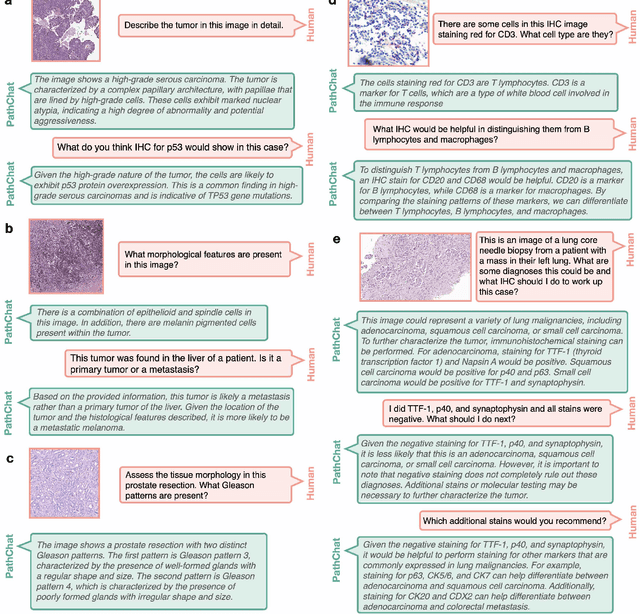
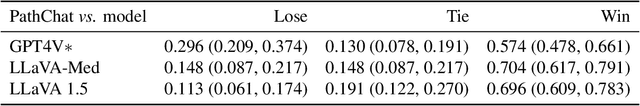
Abstract:The field of computational pathology has witnessed remarkable progress in the development of both task-specific predictive models and task-agnostic self-supervised vision encoders. However, despite the explosive growth of generative artificial intelligence (AI), there has been limited study on building general purpose, multimodal AI assistants tailored to pathology. Here we present PathChat, a vision-language generalist AI assistant for human pathology using an in-house developed foundational vision encoder pretrained on 100 million histology images from over 100,000 patient cases and 1.18 million pathology image-caption pairs. The vision encoder is then combined with a pretrained large language model and the whole system is finetuned on over 250,000 diverse disease agnostic visual language instructions. We compare PathChat against several multimodal vision language AI assistants as well as GPT4V, which powers the commercially available multimodal general purpose AI assistant ChatGPT-4. When relevant clinical context is provided with the histology image, PathChat achieved a diagnostic accuracy of 87% on multiple-choice questions based on publicly available cases of diverse tissue origins and disease models. Additionally, using open-ended questions and human expert evaluation, we found that overall PathChat produced more accurate and pathologist-preferable responses to diverse queries related to pathology. As an interactive and general vision language AI assistant that can flexibly handle both visual and natural language inputs, PathChat can potentially find impactful applications in pathology education, research, and human-in-the-loop clinical decision making.
A General-Purpose Self-Supervised Model for Computational Pathology
Aug 29, 2023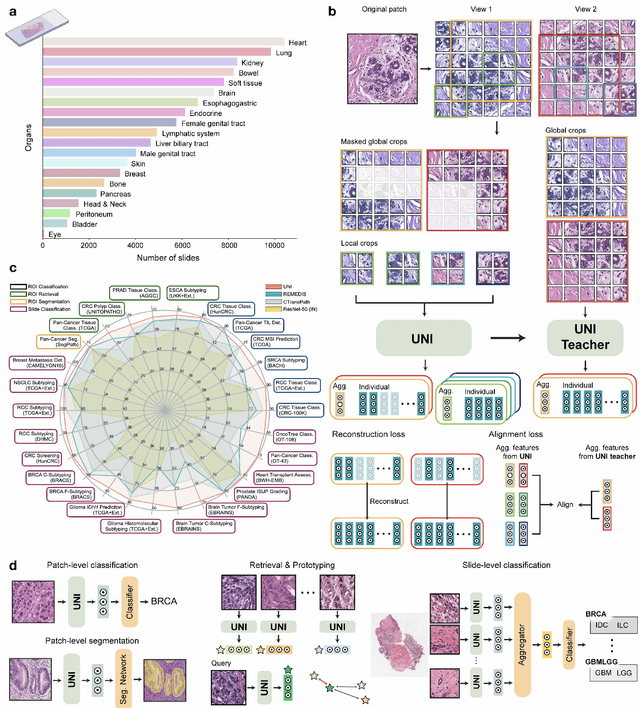

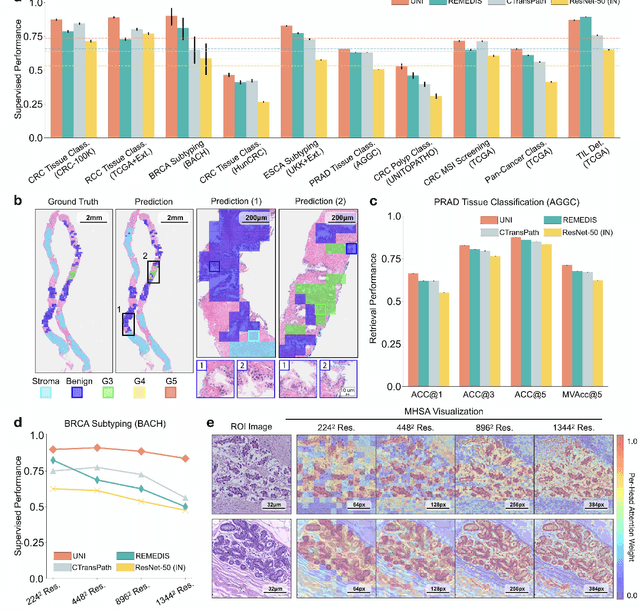

Abstract:Tissue phenotyping is a fundamental computational pathology (CPath) task in learning objective characterizations of histopathologic biomarkers in anatomic pathology. However, whole-slide imaging (WSI) poses a complex computer vision problem in which the large-scale image resolutions of WSIs and the enormous diversity of morphological phenotypes preclude large-scale data annotation. Current efforts have proposed using pretrained image encoders with either transfer learning from natural image datasets or self-supervised pretraining on publicly-available histopathology datasets, but have not been extensively developed and evaluated across diverse tissue types at scale. We introduce UNI, a general-purpose self-supervised model for pathology, pretrained using over 100 million tissue patches from over 100,000 diagnostic haematoxylin and eosin-stained WSIs across 20 major tissue types, and evaluated on 33 representative CPath clinical tasks in CPath of varying diagnostic difficulties. In addition to outperforming previous state-of-the-art models, we demonstrate new modeling capabilities in CPath such as resolution-agnostic tissue classification, slide classification using few-shot class prototypes, and disease subtyping generalization in classifying up to 108 cancer types in the OncoTree code classification system. UNI advances unsupervised representation learning at scale in CPath in terms of both pretraining data and downstream evaluation, enabling data-efficient AI models that can generalize and transfer to a gamut of diagnostically-challenging tasks and clinical workflows in anatomic pathology.
Towards a Visual-Language Foundation Model for Computational Pathology
Jul 25, 2023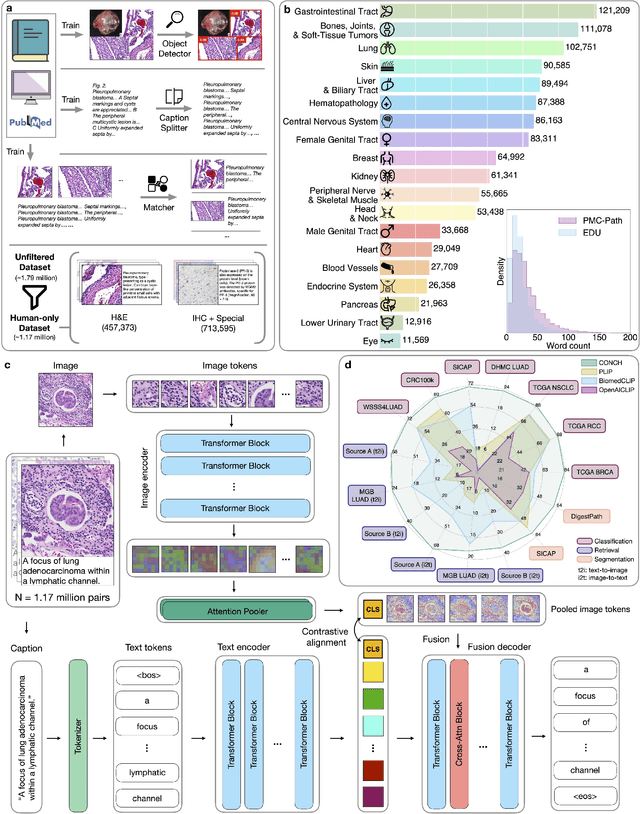
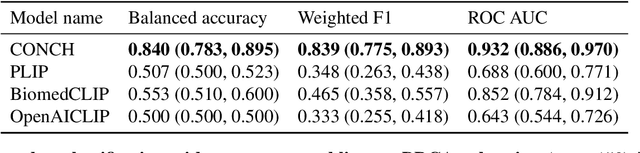
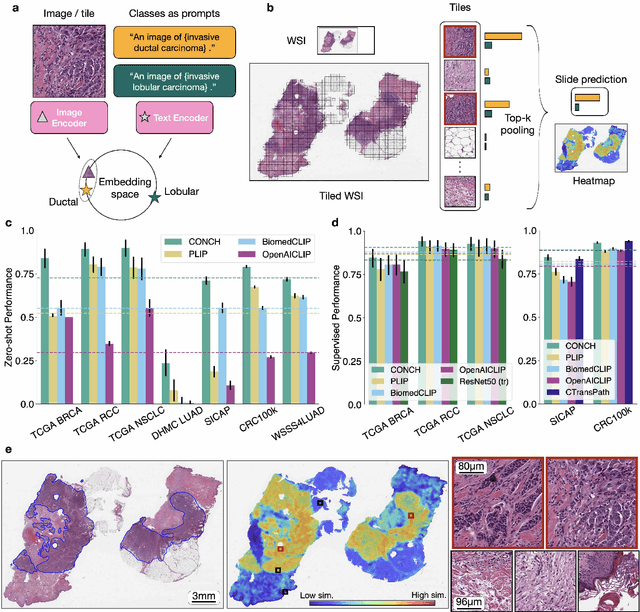

Abstract:The accelerated adoption of digital pathology and advances in deep learning have enabled the development of powerful models for various pathology tasks across a diverse array of diseases and patient cohorts. However, model training is often difficult due to label scarcity in the medical domain and the model's usage is limited by the specific task and disease for which it is trained. Additionally, most models in histopathology leverage only image data, a stark contrast to how humans teach each other and reason about histopathologic entities. We introduce CONtrastive learning from Captions for Histopathology (CONCH), a visual-language foundation model developed using diverse sources of histopathology images, biomedical text, and notably over 1.17 million image-caption pairs via task-agnostic pretraining. Evaluated on a suite of 13 diverse benchmarks, CONCH can be transferred to a wide range of downstream tasks involving either or both histopathology images and text, achieving state-of-the-art performance on histology image classification, segmentation, captioning, text-to-image and image-to-text retrieval. CONCH represents a substantial leap over concurrent visual-language pretrained systems for histopathology, with the potential to directly facilitate a wide array of machine learning-based workflows requiring minimal or no further supervised fine-tuning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge