Faisal Mahmood
NOVA: An Agentic Framework for Automated Histopathology Analysis and Discovery
Nov 14, 2025



Abstract:Digitized histopathology analysis involves complex, time-intensive workflows and specialized expertise, limiting its accessibility. We introduce NOVA, an agentic framework that translates scientific queries into executable analysis pipelines by iteratively generating and running Python code. NOVA integrates 49 domain-specific tools (e.g., nuclei segmentation, whole-slide encoding) built on open-source software, and can also create new tools ad hoc. To evaluate such systems, we present SlideQuest, a 90-question benchmark -- verified by pathologists and biomedical scientists -- spanning data processing, quantitative analysis, and hypothesis testing. Unlike prior biomedical benchmarks focused on knowledge recall or diagnostic QA, SlideQuest demands multi-step reasoning, iterative coding, and computational problem solving. Quantitative evaluation shows NOVA outperforms coding-agent baselines, and a pathologist-verified case study links morphology to prognostically relevant PAM50 subtypes, demonstrating its scalable discovery potential.
Evidence-based diagnostic reasoning with multi-agent copilot for human pathology
Jun 26, 2025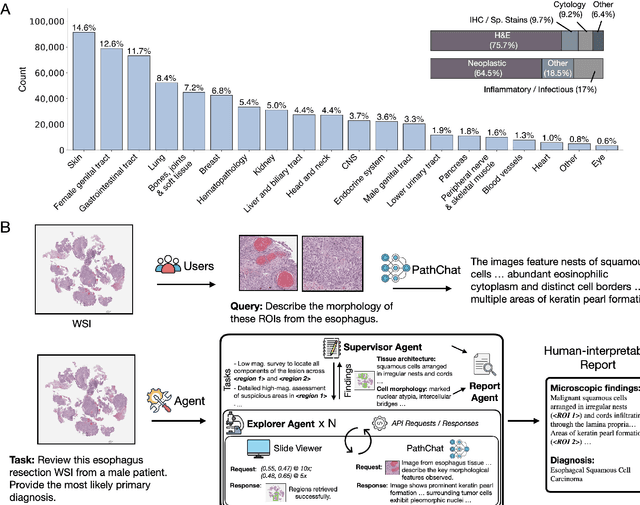
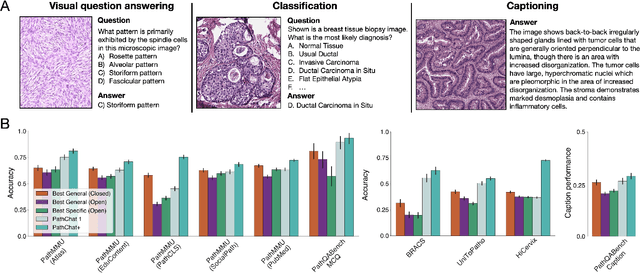
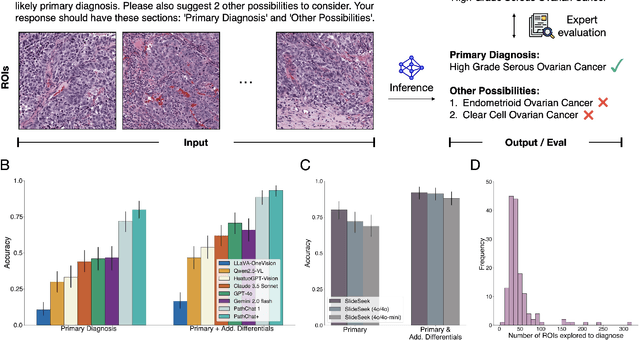
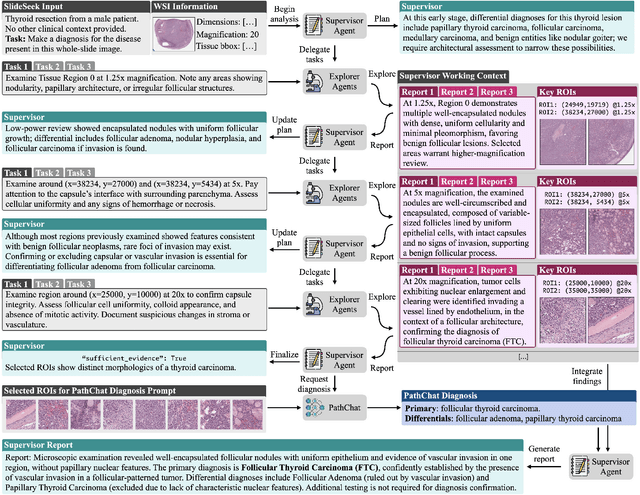
Abstract:Pathology is experiencing rapid digital transformation driven by whole-slide imaging and artificial intelligence (AI). While deep learning-based computational pathology has achieved notable success, traditional models primarily focus on image analysis without integrating natural language instruction or rich, text-based context. Current multimodal large language models (MLLMs) in computational pathology face limitations, including insufficient training data, inadequate support and evaluation for multi-image understanding, and a lack of autonomous, diagnostic reasoning capabilities. To address these limitations, we introduce PathChat+, a new MLLM specifically designed for human pathology, trained on over 1 million diverse, pathology-specific instruction samples and nearly 5.5 million question answer turns. Extensive evaluations across diverse pathology benchmarks demonstrated that PathChat+ substantially outperforms the prior PathChat copilot, as well as both state-of-the-art (SOTA) general-purpose and other pathology-specific models. Furthermore, we present SlideSeek, a reasoning-enabled multi-agent AI system leveraging PathChat+ to autonomously evaluate gigapixel whole-slide images (WSIs) through iterative, hierarchical diagnostic reasoning, reaching high accuracy on DDxBench, a challenging open-ended differential diagnosis benchmark, while also capable of generating visually grounded, humanly-interpretable summary reports.
Do Multiple Instance Learning Models Transfer?
Jun 11, 2025Abstract:Multiple Instance Learning (MIL) is a cornerstone approach in computational pathology (CPath) for generating clinically meaningful slide-level embeddings from gigapixel tissue images. However, MIL often struggles with small, weakly supervised clinical datasets. In contrast to fields such as NLP and conventional computer vision, where transfer learning is widely used to address data scarcity, the transferability of MIL models remains poorly understood. In this study, we systematically evaluate the transfer learning capabilities of pretrained MIL models by assessing 11 models across 21 pretraining tasks for morphological and molecular subtype prediction. Our results show that pretrained MIL models, even when trained on different organs than the target task, consistently outperform models trained from scratch. Moreover, pretraining on pancancer datasets enables strong generalization across organs and tasks, outperforming slide foundation models while using substantially less pretraining data. These findings highlight the robust adaptability of MIL models and demonstrate the benefits of leveraging transfer learning to boost performance in CPath. Lastly, we provide a resource which standardizes the implementation of MIL models and collection of pretrained model weights on popular CPath tasks, available at https://github.com/mahmoodlab/MIL-Lab
AI-driven 3D Spatial Transcriptomics
Feb 25, 2025



Abstract:A comprehensive three-dimensional (3D) map of tissue architecture and gene expression is crucial for illuminating the complexity and heterogeneity of tissues across diverse biomedical applications. However, most spatial transcriptomics (ST) approaches remain limited to two-dimensional (2D) sections of tissue. Although current 3D ST methods hold promise, they typically require extensive tissue sectioning, are complex, are not compatible with non-destructive 3D tissue imaging technologies, and often lack scalability. Here, we present VOlumetrically Resolved Transcriptomics EXpression (VORTEX), an AI framework that leverages 3D tissue morphology and minimal 2D ST to predict volumetric 3D ST. By pretraining on diverse 3D morphology-transcriptomic pairs from heterogeneous tissue samples and then fine-tuning on minimal 2D ST data from a specific volume of interest, VORTEX learns both generic tissue-related and sample-specific morphological correlates of gene expression. This approach enables dense, high-throughput, and fast 3D ST, scaling seamlessly to large tissue volumes far beyond the reach of existing 3D ST techniques. By offering a cost-effective and minimally destructive route to obtaining volumetric molecular insights, we anticipate that VORTEX will accelerate biomarker discovery and our understanding of morphomolecular associations and cell states in complex tissues. Interactive 3D ST volumes can be viewed at https://vortex-demo.github.io/
Accelerating Data Processing and Benchmarking of AI Models for Pathology
Feb 10, 2025Abstract:Advances in foundation modeling have reshaped computational pathology. However, the increasing number of available models and lack of standardized benchmarks make it increasingly complex to assess their strengths, limitations, and potential for further development. To address these challenges, we introduce a new suite of software tools for whole-slide image processing, foundation model benchmarking, and curated publicly available tasks. We anticipate that these resources will promote transparency, reproducibility, and continued progress in the field.
Molecular-driven Foundation Model for Oncologic Pathology
Jan 28, 2025Abstract:Foundation models are reshaping computational pathology by enabling transfer learning, where models pre-trained on vast datasets can be adapted for downstream diagnostic, prognostic, and therapeutic response tasks. Despite these advances, foundation models are still limited in their ability to encode the entire gigapixel whole-slide images without additional training and often lack complementary multimodal data. Here, we introduce Threads, a slide-level foundation model capable of generating universal representations of whole-slide images of any size. Threads was pre-trained using a multimodal learning approach on a diverse cohort of 47,171 hematoxylin and eosin (H&E)-stained tissue sections, paired with corresponding genomic and transcriptomic profiles - the largest such paired dataset to be used for foundation model development to date. This unique training paradigm enables Threads to capture the tissue's underlying molecular composition, yielding powerful representations applicable to a wide array of downstream tasks. In extensive benchmarking across 54 oncology tasks, including clinical subtyping, grading, mutation prediction, immunohistochemistry status determination, treatment response prediction, and survival prediction, Threads outperformed all baselines while demonstrating remarkable generalizability and label efficiency. It is particularly well suited for predicting rare events, further emphasizing its clinical utility. We intend to make the model publicly available for the broader community.
Multimodal Whole Slide Foundation Model for Pathology
Nov 29, 2024



Abstract:The field of computational pathology has been transformed with recent advances in foundation models that encode histopathology region-of-interests (ROIs) into versatile and transferable feature representations via self-supervised learning (SSL). However, translating these advancements to address complex clinical challenges at the patient and slide level remains constrained by limited clinical data in disease-specific cohorts, especially for rare clinical conditions. We propose TITAN, a multimodal whole slide foundation model pretrained using 335,645 WSIs via visual self-supervised learning and vision-language alignment with corresponding pathology reports and 423,122 synthetic captions generated from a multimodal generative AI copilot for pathology. Without any finetuning or requiring clinical labels, TITAN can extract general-purpose slide representations and generate pathology reports that generalize to resource-limited clinical scenarios such as rare disease retrieval and cancer prognosis. We evaluate TITAN on diverse clinical tasks and find that TITAN outperforms both ROI and slide foundation models across machine learning settings such as linear probing, few-shot and zero-shot classification, rare cancer retrieval and cross-modal retrieval, and pathology report generation.
Multistain Pretraining for Slide Representation Learning in Pathology
Aug 05, 2024Abstract:Developing self-supervised learning (SSL) models that can learn universal and transferable representations of H&E gigapixel whole-slide images (WSIs) is becoming increasingly valuable in computational pathology. These models hold the potential to advance critical tasks such as few-shot classification, slide retrieval, and patient stratification. Existing approaches for slide representation learning extend the principles of SSL from small images (e.g., 224 x 224 patches) to entire slides, usually by aligning two different augmentations (or views) of the slide. Yet the resulting representation remains constrained by the limited clinical and biological diversity of the views. Instead, we postulate that slides stained with multiple markers, such as immunohistochemistry, can be used as different views to form a rich task-agnostic training signal. To this end, we introduce Madeleine, a multimodal pretraining strategy for slide representation learning. Madeleine is trained with a dual global-local cross-stain alignment objective on large cohorts of breast cancer samples (N=4,211 WSIs across five stains) and kidney transplant samples (N=12,070 WSIs across four stains). We demonstrate the quality of slide representations learned by Madeleine on various downstream evaluations, ranging from morphological and molecular classification to prognostic prediction, comprising 21 tasks using 7,299 WSIs from multiple medical centers. Code is available at https://github.com/mahmoodlab/MADELEINE.
Composable Interventions for Language Models
Jul 09, 2024



Abstract:Test-time interventions for language models can enhance factual accuracy, mitigate harmful outputs, and improve model efficiency without costly retraining. But despite a flood of new methods, different types of interventions are largely developing independently. In practice, multiple interventions must be applied sequentially to the same model, yet we lack standardized ways to study how interventions interact. We fill this gap by introducing composable interventions, a framework to study the effects of using multiple interventions on the same language models, featuring new metrics and a unified codebase. Using our framework, we conduct extensive experiments and compose popular methods from three emerging intervention categories -- Knowledge Editing, Model Compression, and Machine Unlearning. Our results from 310 different compositions uncover meaningful interactions: compression hinders editing and unlearning, composing interventions hinges on their order of application, and popular general-purpose metrics are inadequate for assessing composability. Taken together, our findings showcase clear gaps in composability, suggesting a need for new multi-objective interventions. All of our code is public: https://github.com/hartvigsen-group/composable-interventions.
Multimodal Prototyping for cancer survival prediction
Jun 28, 2024



Abstract:Multimodal survival methods combining gigapixel histology whole-slide images (WSIs) and transcriptomic profiles are particularly promising for patient prognostication and stratification. Current approaches involve tokenizing the WSIs into smaller patches (>10,000 patches) and transcriptomics into gene groups, which are then integrated using a Transformer for predicting outcomes. However, this process generates many tokens, which leads to high memory requirements for computing attention and complicates post-hoc interpretability analyses. Instead, we hypothesize that we can: (1) effectively summarize the morphological content of a WSI by condensing its constituting tokens using morphological prototypes, achieving more than 300x compression; and (2) accurately characterize cellular functions by encoding the transcriptomic profile with biological pathway prototypes, all in an unsupervised fashion. The resulting multimodal tokens are then processed by a fusion network, either with a Transformer or an optimal transport cross-alignment, which now operates with a small and fixed number of tokens without approximations. Extensive evaluation on six cancer types shows that our framework outperforms state-of-the-art methods with much less computation while unlocking new interpretability analyses.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge