Manuel Tran
B-Cos Aligned Transformers Learn Human-Interpretable Features
Jan 18, 2024Abstract:Vision Transformers (ViTs) and Swin Transformers (Swin) are currently state-of-the-art in computational pathology. However, domain experts are still reluctant to use these models due to their lack of interpretability. This is not surprising, as critical decisions need to be transparent and understandable. The most common approach to understanding transformers is to visualize their attention. However, attention maps of ViTs are often fragmented, leading to unsatisfactory explanations. Here, we introduce a novel architecture called the B-cos Vision Transformer (BvT) that is designed to be more interpretable. It replaces all linear transformations with the B-cos transform to promote weight-input alignment. In a blinded study, medical experts clearly ranked BvTs above ViTs, suggesting that our network is better at capturing biomedically relevant structures. This is also true for the B-cos Swin Transformer (Bwin). Compared to the Swin Transformer, it even improves the F1-score by up to 4.7% on two public datasets.
Training Transitive and Commutative Multimodal Transformers with LoReTTa
May 23, 2023Abstract:Collecting a multimodal dataset with two paired modalities A and B or B and C is difficult in practice. Obtaining a dataset with three aligned modalities A, B, and C is even more challenging. For example, some public medical datasets have only genetic sequences and microscopic images for one patient, and only genetic sequences and radiological images for another - but no dataset includes both microscopic and radiological images for the same patient. This makes it difficult to integrate and combine all modalities into a large pre-trained neural network. We introduce LoReTTa (Linking mOdalities with a tRansitive and commutativE pre-Training sTrAtegy) to address this understudied problem. Our self-supervised framework combines causal masked modeling with the rules of commutativity and transitivity to transition within and between different modalities. Thus, it can model the relation A -> C with A -> B -> C. Given a dataset containing only the disjoint combinations (A, B) and (B, C), we show that a transformer pre-trained with LoReTTa can handle any modality combination at inference time, including the never-seen pair (A, C) and the triplet (A, B, C). We evaluate our approach on a multimodal dataset derived from MNIST containing speech, vision, and language, as well as a real-world medical dataset containing mRNA, miRNA, and RPPA samples from TCGA. Compared to traditional pre-training methods, we observe up to a 100-point reduction in perplexity for autoregressive generation tasks and up to a 15% improvement in classification accuracy for previously unseen modality pairs during the pre-training phase.
S5CL: Unifying Fully-Supervised, Self-Supervised, and Semi-Supervised Learning Through Hierarchical Contrastive Learning
Mar 14, 2022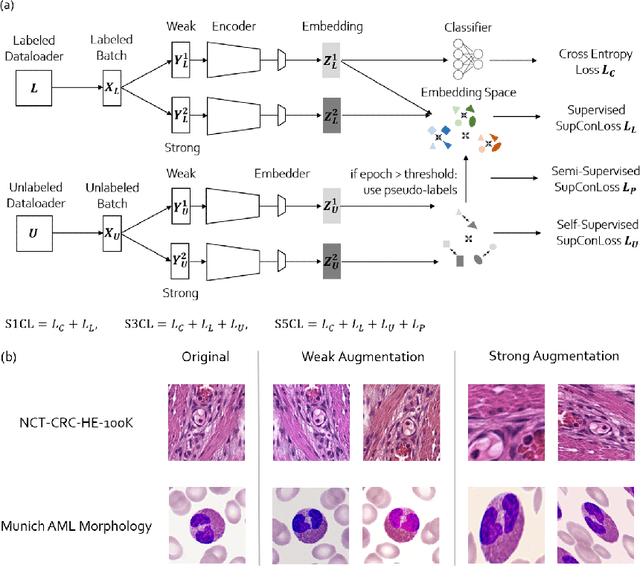
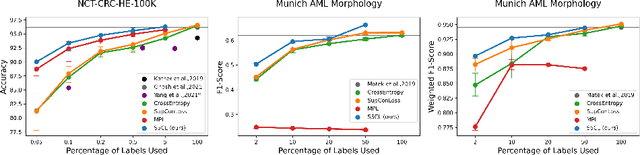
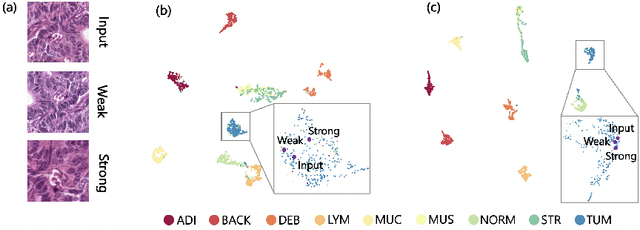
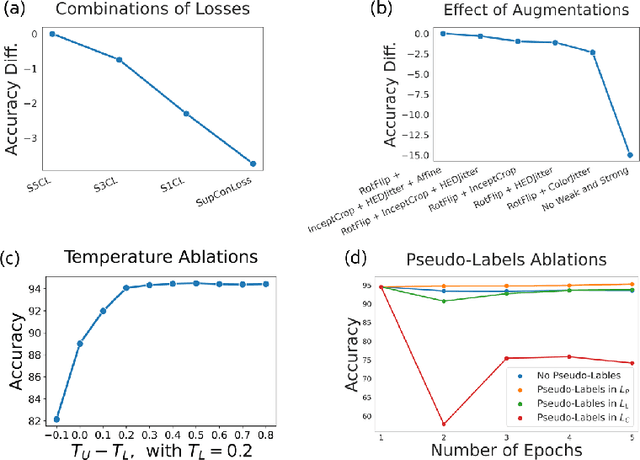
Abstract:In computational pathology, we often face a scarcity of annotations and a large amount of unlabeled data. One method for dealing with this is semi-supervised learning which is commonly split into a self-supervised pretext task and a subsequent model fine-tuning. Here, we compress this two-stage training into one by introducing S5CL, a unified framework for fully-supervised, self-supervised, and semi-supervised learning. With three contrastive losses defined for labeled, unlabeled, and pseudo-labeled images, S5CL can learn feature representations that reflect the hierarchy of distance relationships: similar images and augmentations are embedded the closest, followed by different looking images of the same class, while images from separate classes have the largest distance. Moreover, S5CL allows us to flexibly combine these losses to adapt to different scenarios. Evaluations of our framework on two public histopathological datasets show strong improvements in the case of sparse labels: for a H&E-stained colorectal cancer dataset, the accuracy increases by up to 9% compared to supervised cross-entropy loss; for a highly imbalanced dataset of single white blood cells from leukemia patient blood smears, the F1-score increases by up to 6%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge