Jeroen Hendrikse
Department of Radiology, University Medical Center Utrecht, Utrecht, The Netherlands
A Quantitative Comparison of Epistemic Uncertainty Maps Applied to Multi-Class Segmentation
Sep 22, 2021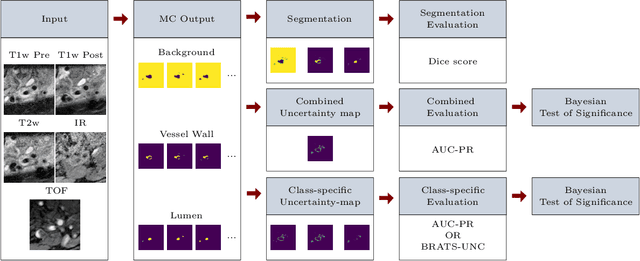
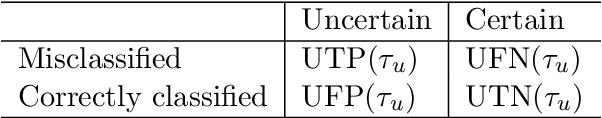
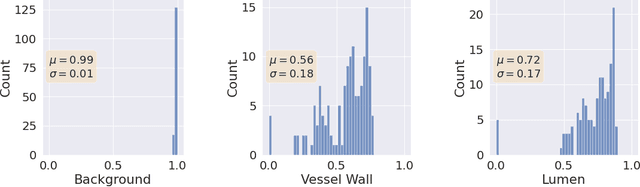
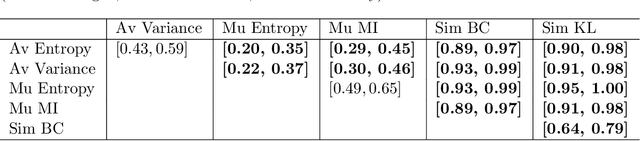
Abstract:Uncertainty assessment has gained rapid interest in medical image analysis. A popular technique to compute epistemic uncertainty is the Monte-Carlo (MC) dropout technique. From a network with MC dropout and a single input, multiple outputs can be sampled. Various methods can be used to obtain epistemic uncertainty maps from those multiple outputs. In the case of multi-class segmentation, the number of methods is even larger as epistemic uncertainty can be computed voxelwise per class or voxelwise per image. This paper highlights a systematic approach to define and quantitatively compare those methods in two different contexts: class-specific epistemic uncertainty maps (one value per image, voxel and class) and combined epistemic uncertainty maps (one value per image and voxel). We applied this quantitative analysis to a multi-class segmentation of the carotid artery lumen and vessel wall, on a multi-center, multi-scanner, multi-sequence dataset of (MR) images. We validated our analysis over 144 sets of hyperparameters of a model. Our main analysis considers the relationship between the order of the voxels sorted according to their epistemic uncertainty values and the misclassification of the prediction. Under this consideration, the comparison of combined uncertainty maps reveals that the multi-class entropy and the multi-class mutual information statistically out-perform the other combined uncertainty maps under study. In a class-specific scenario, the one-versus-all entropy statistically out-performs the class-wise entropy, the class-wise variance and the one versus all mutual information. The class-wise entropy statistically out-performs the other class-specific uncertainty maps in terms of calibration. We made a python package available to reproduce our analysis on different data and tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge