Mohamed S. Elmahdy
CMRINet: Joint Groupwise Registration and Segmentation for Cardiac Function Quantification from Cine-MRI
May 22, 2025Abstract:Accurate and efficient quantification of cardiac function is essential for the estimation of prognosis of cardiovascular diseases (CVDs). One of the most commonly used metrics for evaluating cardiac pumping performance is left ventricular ejection fraction (LVEF). However, LVEF can be affected by factors such as inter-observer variability and varying pre-load and after-load conditions, which can reduce its reproducibility. Additionally, cardiac dysfunction may not always manifest as alterations in LVEF, such as in heart failure and cardiotoxicity diseases. An alternative measure that can provide a relatively load-independent quantitative assessment of myocardial contractility is myocardial strain and strain rate. By using LVEF in combination with myocardial strain, it is possible to obtain a thorough description of cardiac function. Automated estimation of LVEF and other volumetric measures from cine-MRI sequences can be achieved through segmentation models, while strain calculation requires the estimation of tissue displacement between sequential frames, which can be accomplished using registration models. These tasks are often performed separately, potentially limiting the assessment of cardiac function. To address this issue, in this study we propose an end-to-end deep learning (DL) model that jointly estimates groupwise (GW) registration and segmentation for cardiac cine-MRI images. The proposed anatomically-guided Deep GW network was trained and validated on a large dataset of 4-chamber view cine-MRI image series of 374 subjects. A quantitative comparison with conventional GW registration using elastix and two DL-based methods showed that the proposed model improved performance and substantially reduced computation time.
Joint Registration and Segmentation via Multi-Task Learning for Adaptive Radiotherapy of Prostate Cancer
May 05, 2021



Abstract:Medical image registration and segmentation are two of the most frequent tasks in medical image analysis. As these tasks are complementary and correlated, it would be beneficial to apply them simultaneously in a joint manner. In this paper, we formulate registration and segmentation as a joint problem via a Multi-Task Learning (MTL) setting, allowing these tasks to leverage their strengths and mitigate their weaknesses through the sharing of beneficial information. We propose to merge these tasks not only on the loss level, but on the architectural level as well. We studied this approach in the context of adaptive image-guided radiotherapy for prostate cancer, where planning and follow-up CT images as well as their corresponding contours are available for training. The study involves two datasets from different manufacturers and institutes. The first dataset was divided into training (12 patients) and validation (6 patients), and was used to optimize and validate the methodology, while the second dataset (14 patients) was used as an independent test set. We carried out an extensive quantitative comparison between the quality of the automatically generated contours from different network architectures as well as loss weighting methods. Moreover, we evaluated the quality of the generated deformation vector field (DVF). We show that MTL algorithms outperform their Single-Task Learning (STL) counterparts and achieve better generalization on the independent test set. The best algorithm achieved a mean surface distance of $1.06 \pm 0.3$ mm, $1.27 \pm 0.4$ mm, $0.91 \pm 0.4$ mm, and $1.76 \pm 0.8$ mm on the validation set for the prostate, seminal vesicles, bladder, and rectum, respectively. The high accuracy of the proposed method combined with the fast inference speed, makes it a promising method for automatic re-contouring of follow-up scans for adaptive radiotherapy.
Esophageal Tumor Segmentation in CT Images using Dilated Dense Attention Unet (DDAUnet)
Dec 20, 2020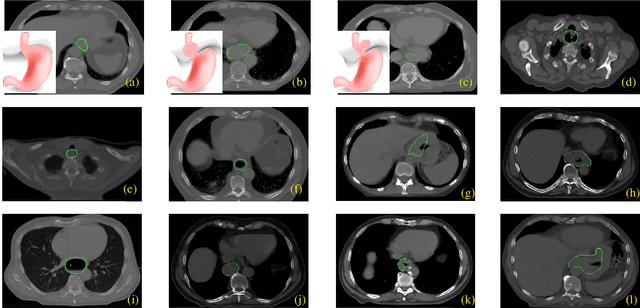
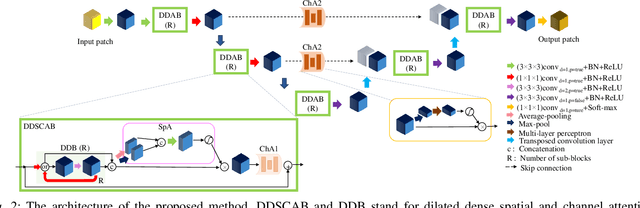
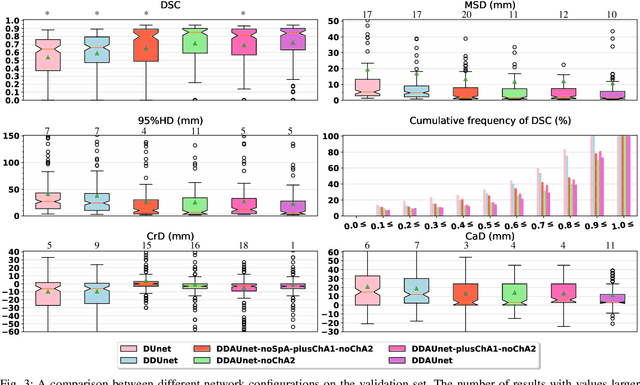

Abstract:Manual or automatic delineation of the esophageal tumor in CT images is known to be very challenging. This is due to the low contrast between the tumor and adjacent tissues, the anatomical variation of the esophagus, as well as the occasional presence of foreign bodies (e.g. feeding tubes). Physicians therefore usually exploit additional knowledge such as endoscopic findings, clinical history, additional imaging modalities like PET scans. Achieving his additional information is time-consuming, while the results are error-prone and might lead to non-deterministic results. In this paper we aim to investigate if and to what extent a simplified clinical workflow based on CT alone, allows one to automatically segment the esophageal tumor with sufficient quality. For this purpose, we present a fully automatic end-to-end esophageal tumor segmentation method based on convolutional neural networks (CNNs). The proposed network, called Dilated Dense Attention Unet (DDAUnet), leverages spatial and channel attention gates in each dense block to selectively concentrate on determinant feature maps and regions. Dilated convolutional layers are used to manage GPU memory and increase the network receptive field. We collected a dataset of 792 scans from 288 distinct patients including varying anatomies with \mbox{air pockets}, feeding tubes and proximal tumors. Repeatability and reproducibility studies were conducted for three distinct splits of training and validation sets. The proposed network achieved a $\mathrm{DSC}$ value of $0.79 \pm 0.20$, a mean surface distance of $5.4 \pm 20.2mm$ and $95\%$ Hausdorff distance of $14.7 \pm 25.0mm$ for 287 test scans, demonstrating promising results with a simplified clinical workflow based on CT alone. Our code is publicly available via \url{https://github.com/yousefis/DenseUnet_Esophagus_Segmentation}.
A Cross-Stitch Architecture for Joint Registration and Segmentation in Adaptive Radiotherapy
Apr 17, 2020



Abstract:Recently, joint registration and segmentation has been formulated in a deep learning setting, by the definition of joint loss functions. In this work, we investigate joining these tasks at the architectural level. We propose a registration network that integrates segmentation propagation between images, and a segmentation network to predict the segmentation directly. These networks are connected into a single joint architecture via so-called cross-stitch units, allowing information to be exchanged between the tasks in a learnable manner. The proposed method is evaluated in the context of adaptive image-guided radiotherapy, using daily prostate CT imaging. Two datasets from different institutes and manufacturers were involved in the study. The first dataset was used for training (12 patients) and validation (6 patients), while the second dataset was used as an independent test set (14 patients). In terms of mean surface distance, our approach achieved $1.06 \pm 0.3$ mm, $0.91 \pm 0.4$ mm, $1.27 \pm 0.4$ mm, and $1.76 \pm 0.8$ mm on the validation set and $1.82 \pm 2.4$ mm, $2.45 \pm 2.4$ mm, $2.45 \pm 5.0$ mm, and $2.57 \pm 2.3$ mm on the test set for the prostate, bladder, seminal vesicles, and rectum, respectively. The proposed multi-task network outperformed single-task networks, as well as a network only joined through the loss function, thus demonstrating the capability to leverage the individual strengths of the segmentation and registration tasks. The obtained performance as well as the inference speed make this a promising candidate for daily re-contouring in adaptive radiotherapy, potentially reducing treatment-related side effects and improving quality-of-life after treatment.
An Adaptive Intelligence Algorithm for Undersampled Knee MRI Reconstruction: Application to the 2019 fastMRI Challenge
Apr 15, 2020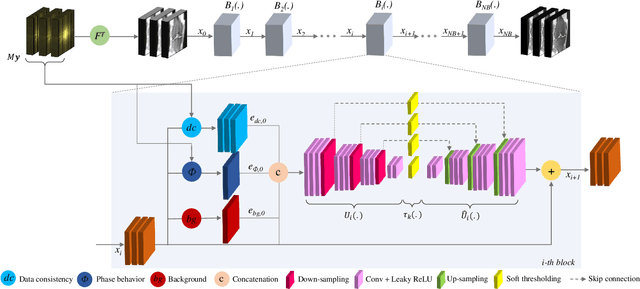
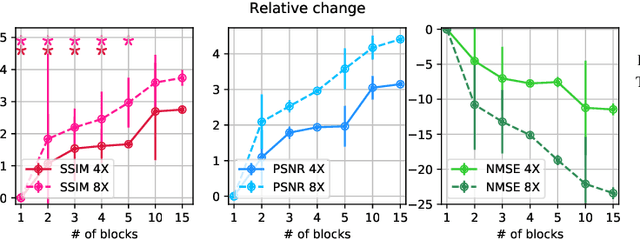
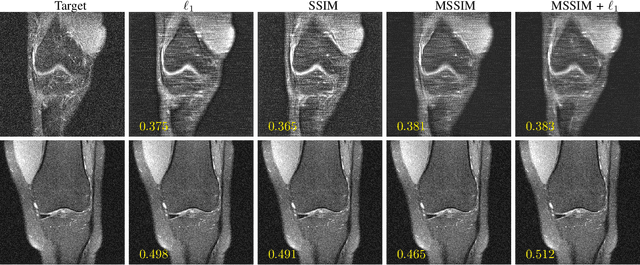

Abstract:Adaptive intelligence aims at empowering machine learning techniques with the additional use of domain knowledge. In this work, we present the application of adaptive intelligence to accelerate MR acquisition. Starting from undersampled k-space data, an iterative learning-based reconstruction scheme inspired by compressed sensing theory is used to reconstruct the images. We adopt deep neural networks to refine and correct prior reconstruction assumptions given the training data. The network was trained and tested on a knee MRI dataset from the 2019 fastMRI challenge organized by Facebook AI Research and NYU Langone Health. All submissions to the challenge were initially ranked based on similarity with a known groundtruth, after which the top 4 submissions were evaluated radiologically. Our method was evaluated by the fastMRI organizers on an independent challenge dataset. It ranked #1, shared #1, and #3 on respectively the 8x accelerated multi-coil, the 4x multi-coil, and the 4x single-coil track. This demonstrates the superior performance and wide applicability of the method.
Patient-Specific Finetuning of Deep Learning Models for Adaptive Radiotherapy in Prostate CT
Feb 17, 2020



Abstract:Contouring of the target volume and Organs-At-Risk (OARs) is a crucial step in radiotherapy treatment planning. In an adaptive radiotherapy setting, updated contours need to be generated based on daily imaging. In this work, we leverage personalized anatomical knowledge accumulated over the treatment sessions, to improve the segmentation accuracy of a pre-trained Convolution Neural Network (CNN), for a specific patient. We investigate a transfer learning approach, fine-tuning the baseline CNN model to a specific patient, based on imaging acquired in earlier treatment fractions. The baseline CNN model is trained on a prostate CT dataset from one hospital of 379 patients. This model is then fine-tuned and tested on an independent dataset of another hospital of 18 patients, each having 7 to 10 daily CT scans. For the prostate, seminal vesicles, bladder and rectum, the model fine-tuned on each specific patient achieved a Mean Surface Distance (MSD) of $1.64 \pm 0.43$ mm, $2.38 \pm 2.76$ mm, $2.30 \pm 0.96$ mm, and $1.24 \pm 0.89$ mm, respectively, which was significantly better than the baseline model. The proposed personalized model adaptation is therefore very promising for clinical implementation in the context of adaptive radiotherapy of prostate cancer.
Fast Dynamic Perfusion and Angiography Reconstruction using an end-to-end 3D Convolutional Neural Network
Sep 04, 2019



Abstract:Hadamard time-encoded pseudo-continuous arterial spin labeling (te-pCASL) is a signal-to-noise ratio (SNR)-efficient MRI technique for acquiring dynamic pCASL signals that encodes the temporal information into the labeling according to a Hadamard matrix. In the decoding step, the contribution of each sub-bolus can be isolated resulting in dynamic perfusion scans. When acquiring te-ASL both with and without flow-crushing, the ASL-signal in the arteries can be isolated resulting in 4D-angiographic information. However, obtaining multi-timepoint perfusion and angiographic data requires two acquisitions. In this study, we propose a 3D Dense-Unet convolutional neural network with a multi-level loss function for reconstructing multi-timepoint perfusion and angiographic information from an interleaved $50\%$-sampled crushed and $50\%$-sampled non-crushed data, thereby negating the additional scan time. We present a framework to generate dynamic pCASL training and validation data, based on models of the intravascular and extravascular te-pCASL signals. The proposed network achieved SSIM values of $97.3 \pm 1.1$ and $96.2 \pm 11.1$ respectively for 4D perfusion and angiographic data reconstruction for 313 test data-sets.
Adversarial optimization for joint registration and segmentation in prostate CT radiotherapy
Jun 28, 2019



Abstract:Joint image registration and segmentation has long been an active area of research in medical imaging. Here, we reformulate this problem in a deep learning setting using adversarial learning. We consider the case in which fixed and moving images as well as their segmentations are available for training, while segmentations are not available during testing; a common scenario in radiotherapy. The proposed framework consists of a 3D end-to-end generator network that estimates the deformation vector field (DVF) between fixed and moving images in an unsupervised fashion and applies this DVF to the moving image and its segmentation. A discriminator network is trained to evaluate how well the moving image and segmentation align with the fixed image and segmentation. The proposed network was trained and evaluated on follow-up prostate CT scans for image-guided radiotherapy, where the planning CT contours are propagated to the daily CT images using the estimated DVF. A quantitative comparison with conventional registration using \texttt{elastix} showed that the proposed method improved performance and substantially reduced computation time, thus enabling real-time contour propagation necessary for online-adaptive radiotherapy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge