Frank J. W. M. Dankers
Multi-Objective Learning to Predict Pareto Fronts Using Hypervolume Maximization
Feb 08, 2021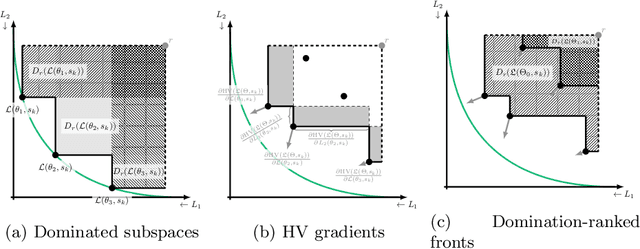
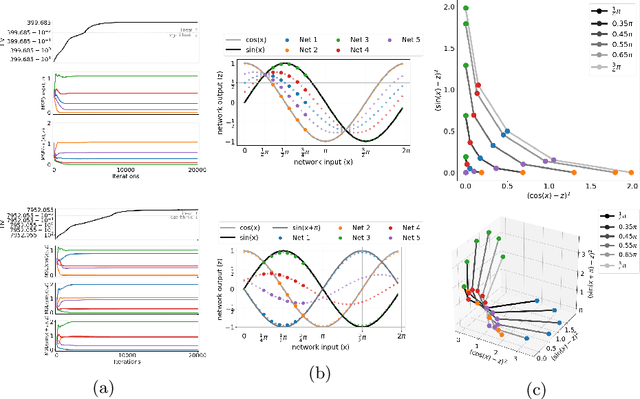
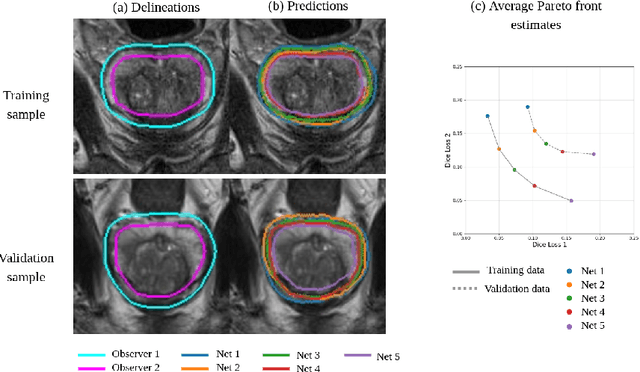
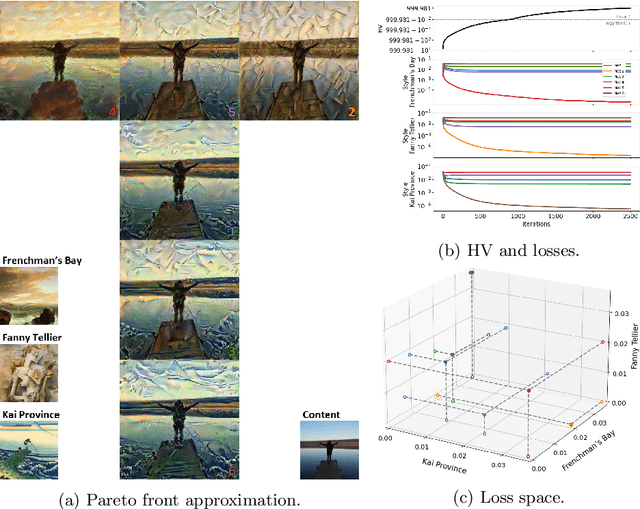
Abstract:Real-world problems are often multi-objective with decision-makers unable to specify a priori which trade-off between the conflicting objectives is preferable. Intuitively, building machine learning solutions in such cases would entail providing multiple predictions that span and uniformly cover the Pareto front of all optimal trade-off solutions. We propose a novel learning approach to estimate the Pareto front by maximizing the dominated hypervolume (HV) of the average loss vectors corresponding to a set of learners, leveraging established multi-objective optimization methods. In our approach, the set of learners are trained multi-objectively with a dynamic loss function, wherein each learner's losses are weighted by their HV maximizing gradients. Consequently, the learners get trained according to different trade-offs on the Pareto front, which otherwise is not guaranteed for fixed linear scalarizations or when optimizing for specific trade-offs per learner without knowing the shape of the Pareto front. Experiments on three different multi-objective tasks show that the outputs of the set of learners are indeed well-spread on the Pareto front. Further, the outputs corresponding to validation samples are also found to closely follow the trade-offs that were learned from training samples for our set of benchmark problems.
Esophageal Tumor Segmentation in CT Images using Dilated Dense Attention Unet (DDAUnet)
Dec 20, 2020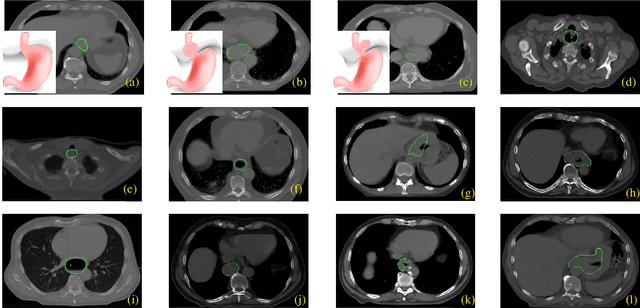
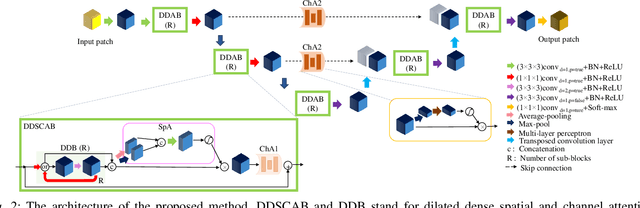
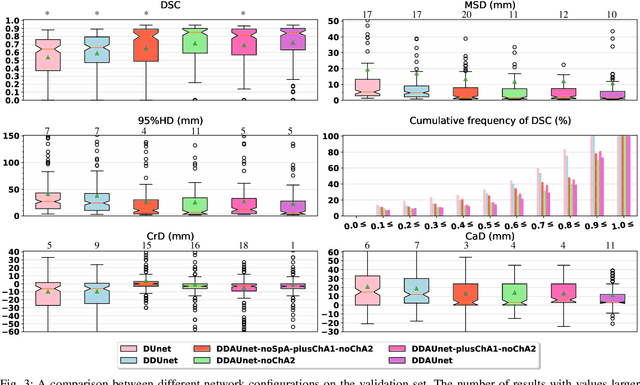

Abstract:Manual or automatic delineation of the esophageal tumor in CT images is known to be very challenging. This is due to the low contrast between the tumor and adjacent tissues, the anatomical variation of the esophagus, as well as the occasional presence of foreign bodies (e.g. feeding tubes). Physicians therefore usually exploit additional knowledge such as endoscopic findings, clinical history, additional imaging modalities like PET scans. Achieving his additional information is time-consuming, while the results are error-prone and might lead to non-deterministic results. In this paper we aim to investigate if and to what extent a simplified clinical workflow based on CT alone, allows one to automatically segment the esophageal tumor with sufficient quality. For this purpose, we present a fully automatic end-to-end esophageal tumor segmentation method based on convolutional neural networks (CNNs). The proposed network, called Dilated Dense Attention Unet (DDAUnet), leverages spatial and channel attention gates in each dense block to selectively concentrate on determinant feature maps and regions. Dilated convolutional layers are used to manage GPU memory and increase the network receptive field. We collected a dataset of 792 scans from 288 distinct patients including varying anatomies with \mbox{air pockets}, feeding tubes and proximal tumors. Repeatability and reproducibility studies were conducted for three distinct splits of training and validation sets. The proposed network achieved a $\mathrm{DSC}$ value of $0.79 \pm 0.20$, a mean surface distance of $5.4 \pm 20.2mm$ and $95\%$ Hausdorff distance of $14.7 \pm 25.0mm$ for 287 test scans, demonstrating promising results with a simplified clinical workflow based on CT alone. Our code is publicly available via \url{https://github.com/yousefis/DenseUnet_Esophagus_Segmentation}.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge