Peter A. N. Bosman
Introns and Templates Matter: Rethinking Linkage in GP-GOMEA
Feb 02, 2026Abstract:GP-GOMEA is among the state-of-the-art for symbolic regression, especially when it comes to finding small and potentially interpretable solutions. A key mechanism employed in any GOMEA variant is the exploitation of linkage, the dependencies between variables, to ensure efficient evolution. In GP-GOMEA, mutual information between node positions in GP trees has so far been used to learn linkage. For this, a fixed expression template is used. This however leads to introns for expressions smaller than the full template. As introns have no impact on fitness, their occurrences are not directly linked to selection. Consequently, introns can adversely affect the extent to which mutual information captures dependencies between tree nodes. To overcome this, we propose two new measures for linkage learning, one that explicitly considers introns in mutual information estimates, and one that revisits linkage learning in GP-GOMEA from a grey-box perspective, yielding a measure that needs not to be learned from the population but is derived directly from the template. Across five standard symbolic regression problems, GP-GOMEA achieves substantial improvements using both measures. We also find that the newly learned linkage structure closely reflects the template linkage structure, and that explicitly using the template structure yields the best performance overall.
Sharing Knowledge without Sharing Data: Stitches can improve ensembles of disjointly trained models
Dec 19, 2025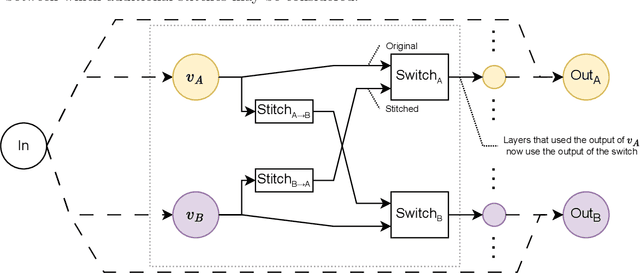
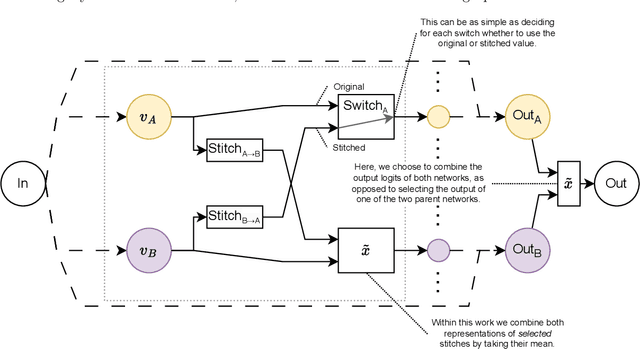
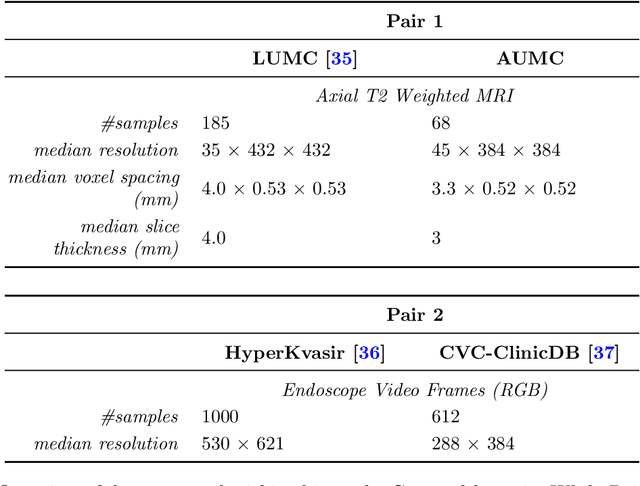
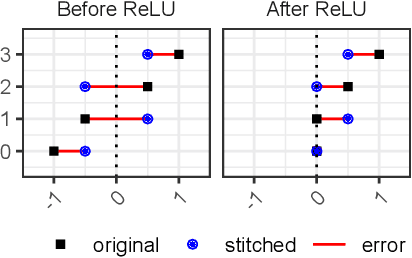
Abstract:Deep learning has been shown to be very capable at performing many real-world tasks. However, this performance is often dependent on the presence of large and varied datasets. In some settings, like in the medical domain, data is often fragmented across parties, and cannot be readily shared. While federated learning addresses this situation, it is a solution that requires synchronicity of parties training a single model together, exchanging information about model weights. We investigate how asynchronous collaboration, where only already trained models are shared (e.g. as part of a publication), affects performance, and propose to use stitching as a method for combining models. Through taking a multi-objective perspective, where performance on each parties' data is viewed independently, we find that training solely on a single parties' data results in similar performance when merging with another parties' data, when considering performance on that single parties' data, while performance on other parties' data is notably worse. Moreover, while an ensemble of such individually trained networks generalizes better, performance on each parties' own dataset suffers. We find that combining intermediate representations in individually trained models with a well placed pair of stitching layers allows this performance to recover to a competitive degree while maintaining improved generalization, showing that asynchronous collaboration can yield competitive results.
Iterated Population Based Training with Task-Agnostic Restarts
Nov 12, 2025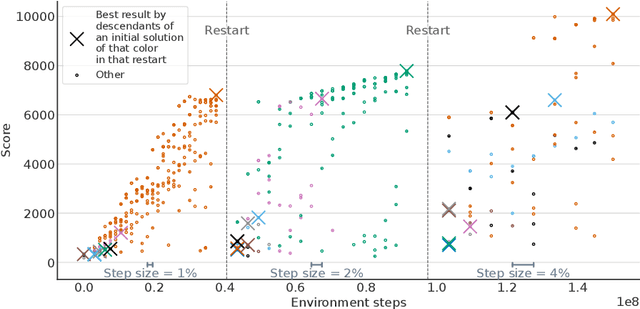
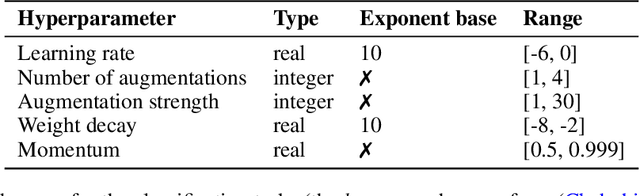
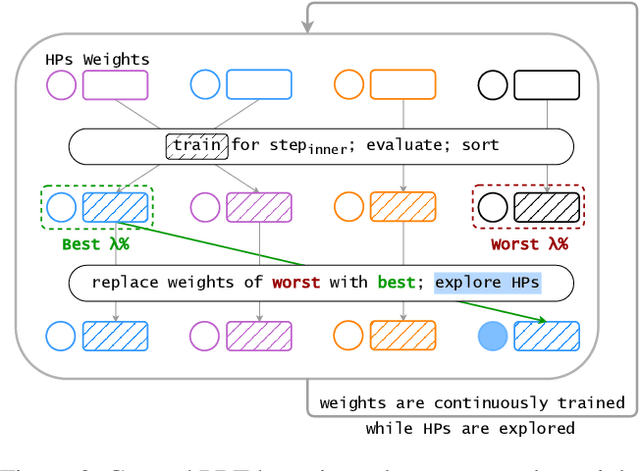
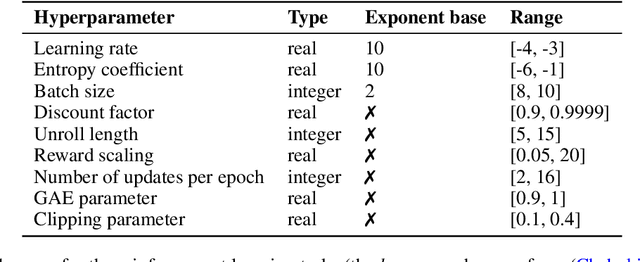
Abstract:Hyperparameter Optimization (HPO) can lift the burden of tuning hyperparameters (HPs) of neural networks. HPO algorithms from the Population Based Training (PBT) family are efficient thanks to dynamically adjusting HPs every few steps of the weight optimization. Recent results indicate that the number of steps between HP updates is an important meta-HP of all PBT variants that can substantially affect their performance. Yet, no method or intuition is available for efficiently setting its value. We introduce Iterated Population Based Training (IPBT), a novel PBT variant that automatically adjusts this HP via restarts that reuse weight information in a task-agnostic way and leverage time-varying Bayesian optimization to reinitialize HPs. Evaluation on 8 image classification and reinforcement learning tasks shows that, on average, our algorithm matches or outperforms 5 previous PBT variants and other HPO algorithms (random search, ASHA, SMAC3), without requiring a budget increase or any changes to its HPs. The source code is available at https://github.com/AwesomeLemon/IPBT.
The Pitfalls and Potentials of Adding Gene-invariance to Optimal Mixing
Jun 18, 2025Abstract:Optimal Mixing (OM) is a variation operator that integrates local search with genetic recombination. EAs with OM are capable of state-of-the-art optimization in discrete spaces, offering significant advantages over classic recombination-based EAs. This success is partly due to high selection pressure that drives rapid convergence. However, this can also negatively impact population diversity, complicating the solving of hierarchical problems, which feature multiple layers of complexity. While there have been attempts to address this issue, these solutions are often complicated and prone to bias. To overcome this, we propose a solution inspired by the Gene Invariant Genetic Algorithm (GIGA), which preserves gene frequencies in the population throughout the process. This technique is tailored to and integrated with the Gene-pool Optimal Mixing Evolutionary Algorithm (GOMEA), resulting in GI-GOMEA. The simple, yet elegant changes are found to have striking potential: GI-GOMEA outperforms GOMEA on a range of well-known problems, even when these problems are adjusted for pitfalls - biases in much-used benchmark problems that can be easily exploited by maintaining gene invariance. Perhaps even more notably, GI-GOMEA is also found to be effective at solving hierarchical problems, including newly introduced asymmetric hierarchical trap functions.
A Step towards Interpretable Multimodal AI Models with MultiFIX
May 16, 2025Abstract:Real-world problems are often dependent on multiple data modalities, making multimodal fusion essential for leveraging diverse information sources. In high-stakes domains, such as in healthcare, understanding how each modality contributes to the prediction is critical to ensure trustworthy and interpretable AI models. We present MultiFIX, an interpretability-driven multimodal data fusion pipeline that explicitly engineers distinct features from different modalities and combines them to make the final prediction. Initially, only deep learning components are used to train a model from data. The black-box (deep learning) components are subsequently either explained using post-hoc methods such as Grad-CAM for images or fully replaced by interpretable blocks, namely symbolic expressions for tabular data, resulting in an explainable model. We study the use of MultiFIX using several training strategies for feature extraction and predictive modeling. Besides highlighting strengths and weaknesses of MultiFIX, experiments on a variety of synthetic datasets with varying degrees of interaction between modalities demonstrate that MultiFIX can generate multimodal models that can be used to accurately explain both the extracted features and their integration without compromising predictive performance.
Thinking Outside the Template with Modular GP-GOMEA
May 02, 2025Abstract:The goal in Symbolic Regression (SR) is to discover expressions that accurately map input to output data. Because often the intent is to understand these expressions, there is a trade-off between accuracy and the interpretability of expressions. GP-GOMEA excels at producing small SR expressions (increasing the potential for interpretability) with high accuracy, but requires a fixed tree template, which limits the types of expressions that can be evolved. This paper presents a modular representation for GP-GOMEA that allows multiple trees to be evolved simultaneously that can be used as (functional) subexpressions. While each tree individually is constrained to a (small) fixed tree template, the final expression, if expanded, can exhibit a much larger structure. Furthermore, the use of subexpressions decomposes the original regression problem and opens the possibility for enhanced interpretability through the piece-wise understanding of small subexpressions. We compare the performance of GP-GOMEA with and without modular templates on a variety of datasets. We find that our proposed approach generally outperforms single-template GP-GOMEA and can moreover uncover ground-truth expressions underlying synthetic datasets with modular subexpressions at a faster rate than GP-GOMEA without modular subexpressions.
Multi-Objective Deep-Learning-based Biomechanical Deformable Image Registration with MOREA
Jan 27, 2025Abstract:When choosing a deformable image registration (DIR) approach for images with large deformations and content mismatch, the realism of found transformations often needs to be traded off against the required runtime. DIR approaches using deep learning (DL) techniques have shown remarkable promise in instantly predicting a transformation. However, on difficult registration problems, the realism of these transformations can fall short. DIR approaches using biomechanical, finite element modeling (FEM) techniques can find more realistic transformations, but tend to require much longer runtimes. This work proposes the first hybrid approach to combine them, with the aim of getting the best of both worlds. This hybrid approach, called DL-MOREA, combines a recently introduced multi-objective DL-based DIR approach which leverages the VoxelMorph framework, called DL-MODIR, with MOREA, an evolutionary algorithm-based, multi-objective DIR approach in which a FEM-like biomechanical mesh transformation model is used. In our proposed hybrid approach, the DL results are used to smartly initialize MOREA, with the aim of more efficiently optimizing its mesh transformation model. We empirically compare DL-MOREA against its components, DL-MODIR and MOREA, on CT scan pairs capturing large bladder filling differences of 15 cervical cancer patients. While MOREA requires a median runtime of 45 minutes, DL-MOREA can already find high-quality transformations after 5 minutes. Compared to the DL-MODIR transformations, the transformations found by DL-MOREA exhibit far less folding and improve or preserve the bladder contour distance error.
Hyperparameter-Free Medical Image Synthesis for Sharing Data and Improving Site-Specific Segmentation
Apr 09, 2024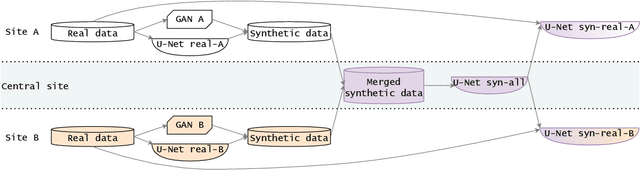
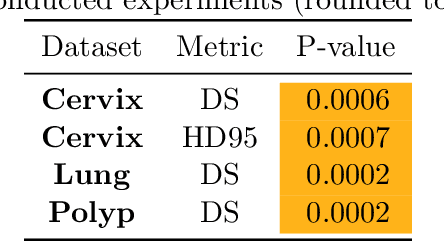
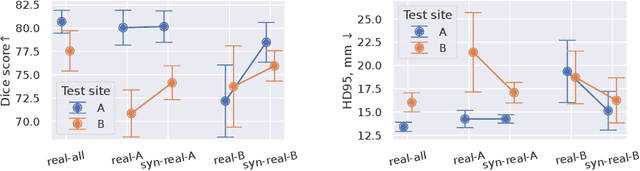
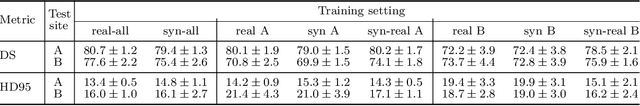
Abstract:Sharing synthetic medical images is a promising alternative to sharing real images that can improve patient privacy and data security. To get good results, existing methods for medical image synthesis must be manually adjusted when they are applied to unseen data. To remove this manual burden, we introduce a Hyperparameter-Free distributed learning method for automatic medical image Synthesis, Sharing, and Segmentation called HyFree-S3. For three diverse segmentation settings (pelvic MRIs, lung X-rays, polyp photos), the use of HyFree-S3 results in improved performance over training only with site-specific data (in the majority of cases). The hyperparameter-free nature of the method should make data synthesis and sharing easier, potentially leading to an increase in the quantity of available data and consequently the quality of the models trained that may ultimately be applied in the clinic. Our code is available at https://github.com/AwesomeLemon/HyFree-S3
Stitching for Neuroevolution: Recombining Deep Neural Networks without Breaking Them
Mar 21, 2024


Abstract:Traditional approaches to neuroevolution often start from scratch. This becomes prohibitively expensive in terms of computational and data requirements when targeting modern, deep neural networks. Using a warm start could be highly advantageous, e.g., using previously trained networks, potentially from different sources. This moreover enables leveraging the benefits of transfer learning (in particular vastly reduced training effort). However, recombining trained networks is non-trivial because architectures and feature representations typically differ. Consequently, a straightforward exchange of layers tends to lead to a performance breakdown. We overcome this by matching the layers of parent networks based on their connectivity, identifying potential crossover points. To correct for differing feature representations between these layers we employ stitching, which merges the networks by introducing new layers at crossover points. To train the merged network, only stitching layers need to be considered. New networks can then be created by selecting a subnetwork by choosing which stitching layers to (not) use. Assessing their performance is efficient as only their evaluation on data is required. We experimentally show that our approach enables finding networks that represent novel trade-offs between performance and computational cost, with some even dominating the original networks.
Multi-Objective Learning for Deformable Image Registration
Feb 23, 2024Abstract:Deformable image registration (DIR) involves optimization of multiple conflicting objectives, however, not many existing DIR algorithms are multi-objective (MO). Further, while there has been progress in the design of deep learning algorithms for DIR, there is no work in the direction of MO DIR using deep learning. In this paper, we fill this gap by combining a recently proposed approach for MO training of neural networks with a well-known deep neural network for DIR and create a deep learning based MO DIR approach. We evaluate the proposed approach for DIR of pelvic magnetic resonance imaging (MRI) scans. We experimentally demonstrate that the proposed MO DIR approach -- providing multiple registration outputs for each patient that each correspond to a different trade-off between the objectives -- has additional desirable properties from a clinical use point-of-view as compared to providing a single DIR output. The experiments also show that the proposed MO DIR approach provides a better spread of DIR outputs across the entire trade-off front than simply training multiple neural networks with weights for each objective sampled from a grid of possible values.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge