Thalea Schlender
A Step towards Interpretable Multimodal AI Models with MultiFIX
May 16, 2025Abstract:Real-world problems are often dependent on multiple data modalities, making multimodal fusion essential for leveraging diverse information sources. In high-stakes domains, such as in healthcare, understanding how each modality contributes to the prediction is critical to ensure trustworthy and interpretable AI models. We present MultiFIX, an interpretability-driven multimodal data fusion pipeline that explicitly engineers distinct features from different modalities and combines them to make the final prediction. Initially, only deep learning components are used to train a model from data. The black-box (deep learning) components are subsequently either explained using post-hoc methods such as Grad-CAM for images or fully replaced by interpretable blocks, namely symbolic expressions for tabular data, resulting in an explainable model. We study the use of MultiFIX using several training strategies for feature extraction and predictive modeling. Besides highlighting strengths and weaknesses of MultiFIX, experiments on a variety of synthetic datasets with varying degrees of interaction between modalities demonstrate that MultiFIX can generate multimodal models that can be used to accurately explain both the extracted features and their integration without compromising predictive performance.
MultiFIX: An XAI-friendly feature inducing approach to building models from multimodal data
Feb 19, 2024Abstract:In the health domain, decisions are often based on different data modalities. Thus, when creating prediction models, multimodal fusion approaches that can extract and combine relevant features from different data modalities, can be highly beneficial. Furthermore, it is important to understand how each modality impacts the final prediction, especially in high-stake domains, so that these models can be used in a trustworthy and responsible manner. We propose MultiFIX: a new interpretability-focused multimodal data fusion pipeline that explicitly induces separate features from different data types that can subsequently be combined to make a final prediction. An end-to-end deep learning architecture is used to train a predictive model and extract representative features of each modality. Each part of the model is then explained using explainable artificial intelligence techniques. Attention maps are used to highlight important regions in image inputs. Inherently interpretable symbolic expressions, learned with GP-GOMEA, are used to describe the contribution of tabular inputs. The fusion of the extracted features to predict the target label is also replaced by a symbolic expression, learned with GP-GOMEA. Results on synthetic problems demonstrate the strengths and limitations of MultiFIX. Lastly, we apply MultiFIX to a publicly available dataset for the detection of malignant skin lesions.
Improving the efficiency of GP-GOMEA for higher-arity operators
Feb 15, 2024Abstract:Deploying machine learning models into sensitive domains in our society requires these models to be explainable. Genetic Programming (GP) can offer a way to evolve inherently interpretable expressions. GP-GOMEA is a form of GP that has been found particularly effective at evolving expressions that are accurate yet of limited size and, thus, promote interpretability. Despite this strength, a limitation of GP-GOMEA is template-based. This negatively affects its scalability regarding the arity of operators that can be used, since with increasing operator arity, an increasingly large part of the template tends to go unused. In this paper, we therefore propose two enhancements to GP-GOMEA: (i) semantic subtree inheritance, which performs additional variation steps that consider the semantic context of a subtree, and (ii) greedy child selection, which explicitly considers parts of the template that in standard GP-GOMEA remain unused. We compare different versions of GP-GOMEA regarding search enhancements on a set of continuous and discontinuous regression problems, with varying tree depths and operator sets. Experimental results show that both proposed search enhancements have a generally positive impact on the performance of GP-GOMEA, especially when the set of operators to choose from is large and contains higher-arity operators.
"Thy algorithm shalt not bear false witness": An Evaluation of Multiclass Debiasing Methods on Word Embeddings
Nov 04, 2020
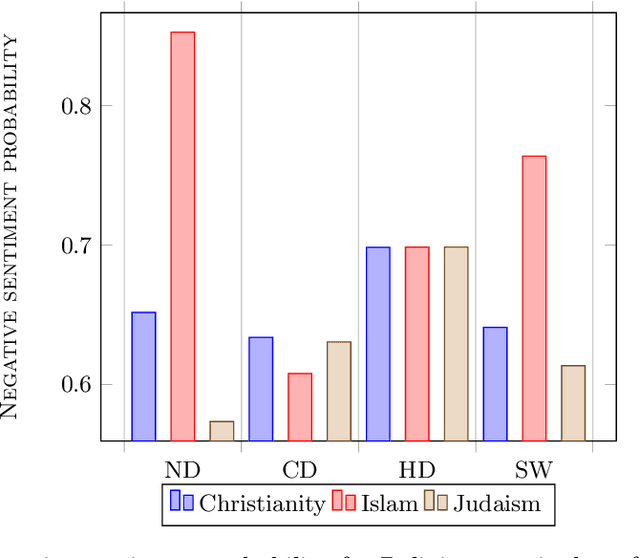
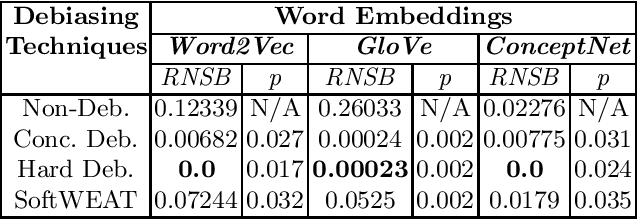
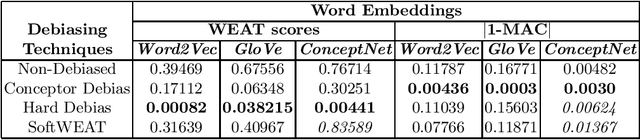
Abstract:With the vast development and employment of artificial intelligence applications, research into the fairness of these algorithms has been increased. Specifically, in the natural language processing domain, it has been shown that social biases persist in word embeddings and are thus in danger of amplifying these biases when used. As an example of social bias, religious biases are shown to persist in word embeddings and the need for its removal is highlighted. This paper investigates the state-of-the-art multiclass debiasing techniques: Hard debiasing, SoftWEAT debiasing and Conceptor debiasing. It evaluates their performance when removing religious bias on a common basis by quantifying bias removal via the Word Embedding Association Test (WEAT), Mean Average Cosine Similarity (MAC) and the Relative Negative Sentiment Bias (RNSB). By investigating the religious bias removal on three widely used word embeddings, namely: Word2Vec, GloVe, and ConceptNet, it is shown that the preferred method is ConceptorDebiasing. Specifically, this technique manages to decrease the measured religious bias on average by 82,42%, 96,78% and 54,76% for the three word embedding sets respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge