Monika Grewal
Multi-Objective Learning for Deformable Image Registration
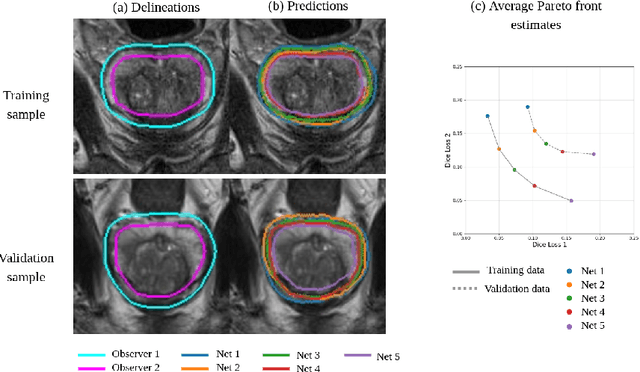
Feb 23, 2024Abstract:Deformable image registration (DIR) involves optimization of multiple conflicting objectives, however, not many existing DIR algorithms are multi-objective (MO). Further, while there has been progress in the design of deep learning algorithms for DIR, there is no work in the direction of MO DIR using deep learning. In this paper, we fill this gap by combining a recently proposed approach for MO training of neural networks with a well-known deep neural network for DIR and create a deep learning based MO DIR approach. We evaluate the proposed approach for DIR of pelvic magnetic resonance imaging (MRI) scans. We experimentally demonstrate that the proposed MO DIR approach -- providing multiple registration outputs for each patient that each correspond to a different trade-off between the objectives -- has additional desirable properties from a clinical use point-of-view as compared to providing a single DIR output. The experiments also show that the proposed MO DIR approach provides a better spread of DIR outputs across the entire trade-off front than simply training multiple neural networks with weights for each objective sampled from a grid of possible values.
Clinically Acceptable Segmentation of Organs at Risk in Cervical Cancer Radiation Treatment from Clinically Available Annotations
Feb 21, 2023Abstract:Deep learning models benefit from training with a large dataset (labeled or unlabeled). Following this motivation, we present an approach to learn a deep learning model for the automatic segmentation of Organs at Risk (OARs) in cervical cancer radiation treatment from a large clinically available dataset of Computed Tomography (CT) scans containing data inhomogeneity, label noise, and missing annotations. We employ simple heuristics for automatic data cleaning to minimize data inhomogeneity and label noise. Further, we develop a semi-supervised learning approach utilizing a teacher-student setup, annotation imputation, and uncertainty-guided training to learn in presence of missing annotations. Our experimental results show that learning from a large dataset with our approach yields a significant improvement in the test performance despite missing annotations in the data. Further, the contours generated from the segmentation masks predicted by our model are found to be equally clinically acceptable as manually generated contours.
Mixed-Block Neural Architecture Search for Medical Image Segmentation
Feb 23, 2022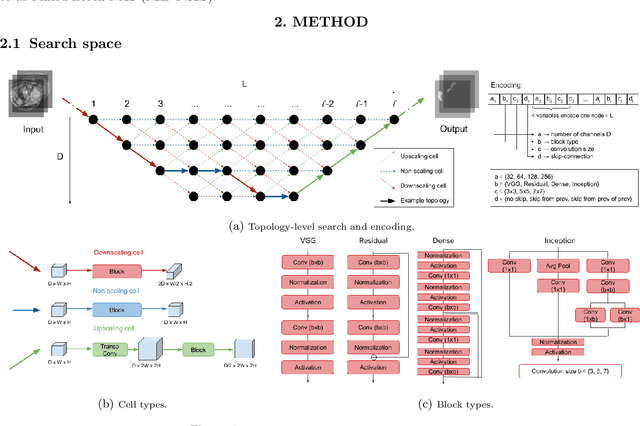
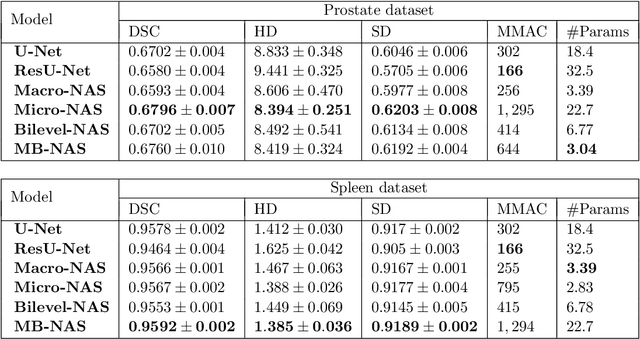
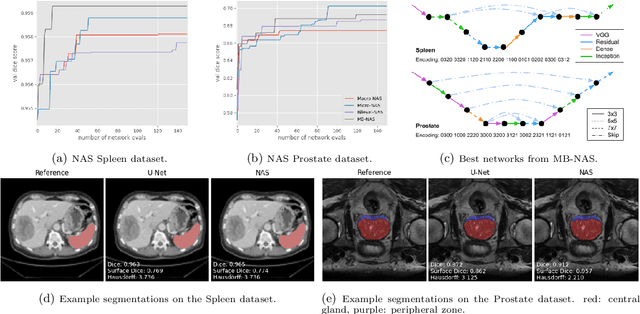
Abstract:Deep Neural Networks (DNNs) have the potential for making various clinical procedures more time-efficient by automating medical image segmentation. Due to their strong, in some cases human-level, performance, they have become the standard approach in this field. The design of the best possible medical image segmentation DNNs, however, is task-specific. Neural Architecture Search (NAS), i.e., the automation of neural network design, has been shown to have the capability to outperform manually designed networks for various tasks. However, the existing NAS methods for medical image segmentation have explored a quite limited range of types of DNN architectures that can be discovered. In this work, we propose a novel NAS search space for medical image segmentation networks. This search space combines the strength of a generalised encoder-decoder structure, well known from U-Net, with network blocks that have proven to have a strong performance in image classification tasks. The search is performed by looking for the best topology of multiple cells simultaneously with the configuration of each cell within, allowing for interactions between topology and cell-level attributes. From experiments on two publicly available datasets, we find that the networks discovered by our proposed NAS method have better performance than well-known handcrafted segmentation networks, and outperform networks found with other NAS approaches that perform only topology search, and topology-level search followed by cell-level search.
Automatic Landmarks Correspondence Detection in Medical Images with an Application to Deformable Image Registration
Sep 06, 2021



Abstract:Deformable Image Registration (DIR) can benefit from additional guidance using corresponding landmarks in the images. However, the benefits thereof are largely understudied, especially due to the lack of automatic detection methods for corresponding landmarks in three-dimensional (3D) medical images. In this work, we present a Deep Convolutional Neural Network (DCNN), called DCNN-Match, that learns to predict landmark correspondences in 3D images in a self-supervised manner. We explored five variants of DCNN-Match that use different loss functions and tested DCNN-Match separately as well as in combination with the open-source registration software Elastix to assess its impact on a common DIR approach. We employed lower-abdominal Computed Tomography (CT) scans from cervical cancer patients: 121 pelvic CT scan pairs containing simulated elastic transformations and 11 pairs demonstrating clinical deformations. Our results show significant improvement in DIR performance when landmark correspondences predicted by DCNN-Match were used in case of simulated as well as clinical deformations. We also observed that the spatial distribution of the automatically identified landmarks and the associated matching errors affect the extent of improvement in DIR. Finally, DCNN-Match was found to generalize well to Magnetic Resonance Imaging (MRI) scans without requiring retraining, indicating easy applicability to other datasets.
Multi-Objective Learning to Predict Pareto Fronts Using Hypervolume Maximization
Feb 08, 2021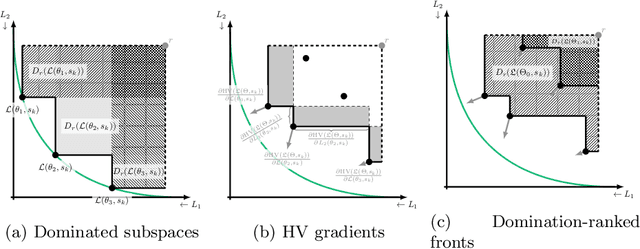
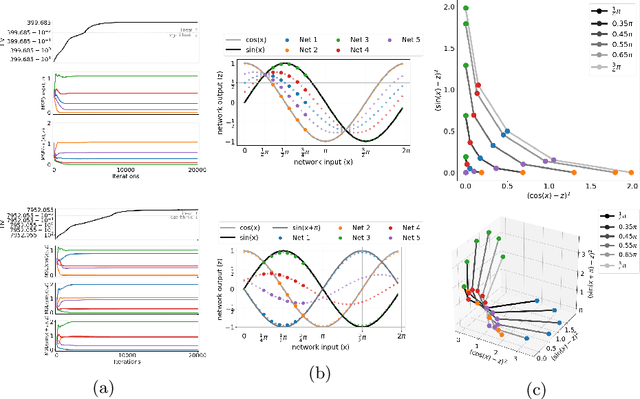
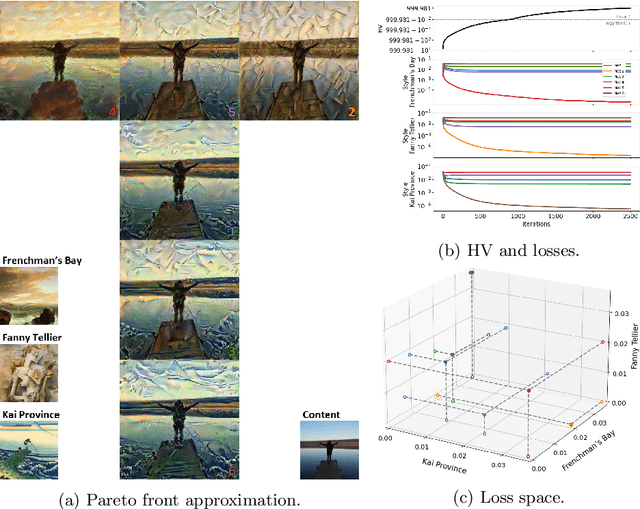
Abstract:Real-world problems are often multi-objective with decision-makers unable to specify a priori which trade-off between the conflicting objectives is preferable. Intuitively, building machine learning solutions in such cases would entail providing multiple predictions that span and uniformly cover the Pareto front of all optimal trade-off solutions. We propose a novel learning approach to estimate the Pareto front by maximizing the dominated hypervolume (HV) of the average loss vectors corresponding to a set of learners, leveraging established multi-objective optimization methods. In our approach, the set of learners are trained multi-objectively with a dynamic loss function, wherein each learner's losses are weighted by their HV maximizing gradients. Consequently, the learners get trained according to different trade-offs on the Pareto front, which otherwise is not guaranteed for fixed linear scalarizations or when optimizing for specific trade-offs per learner without knowing the shape of the Pareto front. Experiments on three different multi-objective tasks show that the outputs of the set of learners are indeed well-spread on the Pareto front. Further, the outputs corresponding to validation samples are also found to closely follow the trade-offs that were learned from training samples for our set of benchmark problems.
An End-to-end Deep Learning Approach for Landmark Detection and Matching in Medical Images
Jan 21, 2020



Abstract:Anatomical landmark correspondences in medical images can provide additional guidance information for the alignment of two images, which, in turn, is crucial for many medical applications. However, manual landmark annotation is labor-intensive. Therefore, we propose an end-to-end deep learning approach to automatically detect landmark correspondences in pairs of two-dimensional (2D) images. Our approach consists of a Siamese neural network, which is trained to identify salient locations in images as landmarks and predict matching probabilities for landmark pairs from two different images. We trained our approach on 2D transverse slices from 168 lower abdominal Computed Tomography (CT) scans. We tested the approach on 22,206 pairs of 2D slices with varying levels of intensity, affine, and elastic transformations. The proposed approach finds an average of 639, 466, and 370 landmark matches per image pair for intensity, affine, and elastic transformations, respectively, with spatial matching errors of at most 1 mm. Further, more than 99% of the landmark pairs are within a spatial matching error of 2 mm, 4 mm, and 8 mm for image pairs with intensity, affine, and elastic transformations, respectively. To investigate the utility of our developed approach in a clinical setting, we also tested our approach on pairs of transverse slices selected from follow-up CT scans of three patients. Visual inspection of the results revealed landmark matches in both bony anatomical regions as well as in soft tissues lacking prominent intensity gradients.
Anatomical labeling of brain CT scan anomalies using multi-context nearest neighbor relation networks
Jan 22, 2018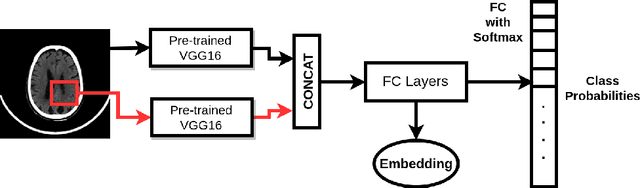
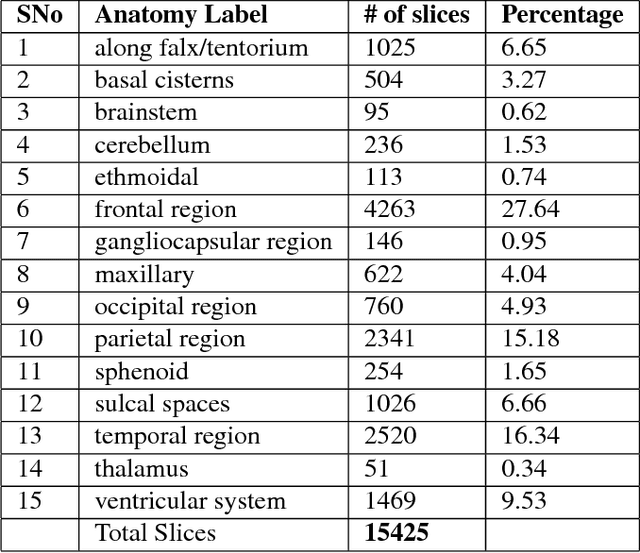
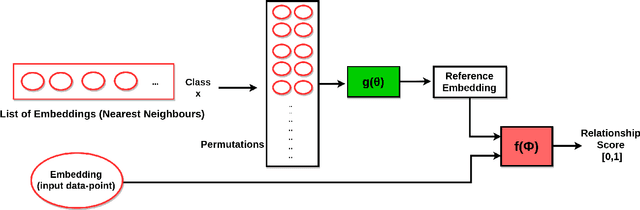
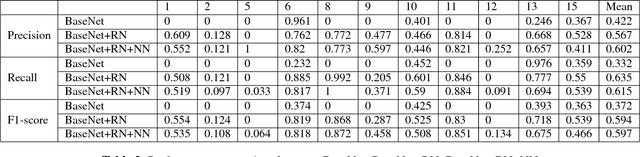
Abstract:This work is an endeavor to develop a deep learning methodology for automated anatomical labeling of a given region of interest (ROI) in brain computed tomography (CT) scans. We combine both local and global context to obtain a representation of the ROI. We then use Relation Networks (RNs) to predict the corresponding anatomy of the ROI based on its relationship score for each class. Further, we propose a novel strategy employing nearest neighbors approach for training RNs. We train RNs to learn the relationship of the target ROI with the joint representation of its nearest neighbors in each class instead of all data-points in each class. The proposed strategy leads to better training of RNs along with increased performance as compared to training baseline RN network.
RADNET: Radiologist Level Accuracy using Deep Learning for HEMORRHAGE detection in CT Scans
Jan 03, 2018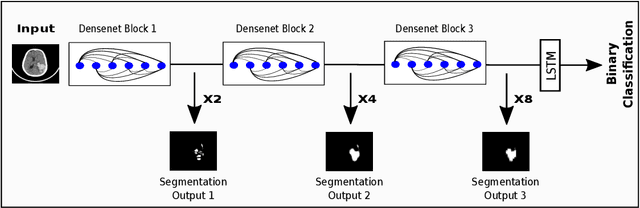
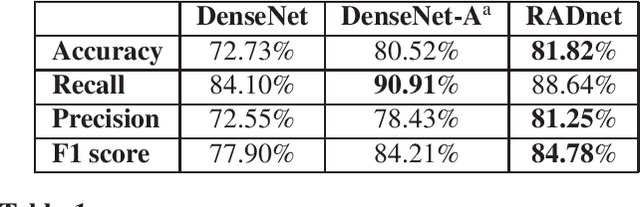
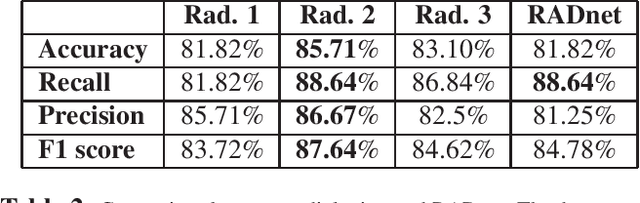
Abstract:We describe a deep learning approach for automated brain hemorrhage detection from computed tomography (CT) scans. Our model emulates the procedure followed by radiologists to analyse a 3D CT scan in real-world. Similar to radiologists, the model sifts through 2D cross-sectional slices while paying close attention to potential hemorrhagic regions. Further, the model utilizes 3D context from neighboring slices to improve predictions at each slice and subsequently, aggregates the slice-level predictions to provide diagnosis at CT level. We refer to our proposed approach as Recurrent Attention DenseNet (RADnet) as it employs original DenseNet architecture along with adding the components of attention for slice level predictions and recurrent neural network layer for incorporating 3D context. The real-world performance of RADnet has been benchmarked against independent analysis performed by three senior radiologists for 77 brain CTs. RADnet demonstrates 81.82% hemorrhage prediction accuracy at CT level that is comparable to radiologists. Further, RADnet achieves higher recall than two of the three radiologists, which is remarkable.
Boosted Cascaded Convnets for Multilabel Classification of Thoracic Diseases in Chest Radiographs
Nov 23, 2017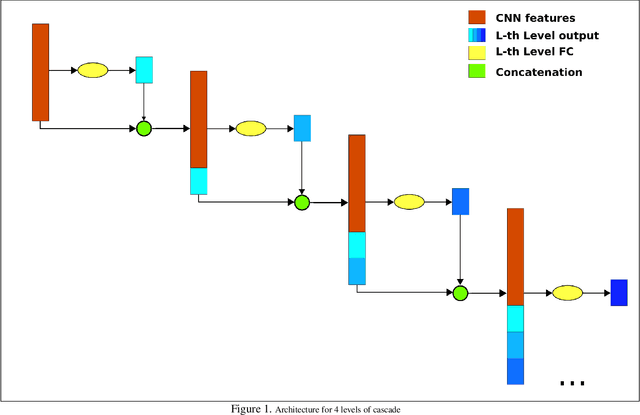
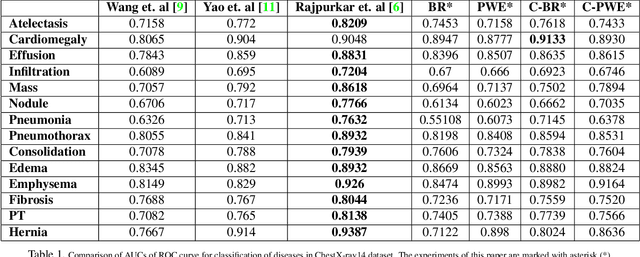
Abstract:Chest X-ray is one of the most accessible medical imaging technique for diagnosis of multiple diseases. With the availability of ChestX-ray14, which is a massive dataset of chest X-ray images and provides annotations for 14 thoracic diseases; it is possible to train Deep Convolutional Neural Networks (DCNN) to build Computer Aided Diagnosis (CAD) systems. In this work, we experiment a set of deep learning models and present a cascaded deep neural network that can diagnose all 14 pathologies better than the baseline and is competitive with other published methods. Our work provides the quantitative results to answer following research questions for the dataset: 1) What loss functions to use for training DCNN from scratch on ChestX-ray14 dataset that demonstrates high class imbalance and label co occurrence? 2) How to use cascading to model label dependency and to improve accuracy of the deep learning model?
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge