Arkadiy Dushatskiy
Solving the Tree Containment Problem Using Graph Neural Networks
Apr 15, 2024Abstract:Tree Containment is a fundamental problem in phylogenetics useful for verifying a proposed phylogenetic network, representing the evolutionary history of certain species. Tree Containment asks whether the given phylogenetic tree (for instance, constructed from a DNA fragment showing tree-like evolution) is contained in the given phylogenetic network. In the general case, this is an NP-complete problem. We propose to solve it approximately using Graph Neural Networks. In particular, we propose to combine the given network and the tree and apply a Graph Neural Network to this network-tree graph. This way, we achieve the capability of solving the tree containment instances representing a larger number of species than the instances contained in the training dataset (i.e., our algorithm has the inductive learning ability). Our algorithm demonstrates an accuracy of over $95\%$ in solving the tree containment problem on instances with up to 100 leaves.
Shrink-Perturb Improves Architecture Mixing during Population Based Training for Neural Architecture Search
Jul 28, 2023Abstract:In this work, we show that simultaneously training and mixing neural networks is a promising way to conduct Neural Architecture Search (NAS). For hyperparameter optimization, reusing the partially trained weights allows for efficient search, as was previously demonstrated by the Population Based Training (PBT) algorithm. We propose PBT-NAS, an adaptation of PBT to NAS where architectures are improved during training by replacing poorly-performing networks in a population with the result of mixing well-performing ones and inheriting the weights using the shrink-perturb technique. After PBT-NAS terminates, the created networks can be directly used without retraining. PBT-NAS is highly parallelizable and effective: on challenging tasks (image generation and reinforcement learning) PBT-NAS achieves superior performance compared to baselines (random search and mutation-based PBT).
Multi-Objective Population Based Training
Jun 02, 2023Abstract:Population Based Training (PBT) is an efficient hyperparameter optimization algorithm. PBT is a single-objective algorithm, but many real-world hyperparameter optimization problems involve two or more conflicting objectives. In this work, we therefore introduce a multi-objective version of PBT, MO-PBT. Our experiments on diverse multi-objective hyperparameter optimization problems (Precision/Recall, Accuracy/Fairness, Accuracy/Adversarial Robustness) show that MO-PBT outperforms random search, single-objective PBT, and the state-of-the-art multi-objective hyperparameter optimization algorithm MO-ASHA.
Data variation-aware medical image segmentation
Feb 24, 2022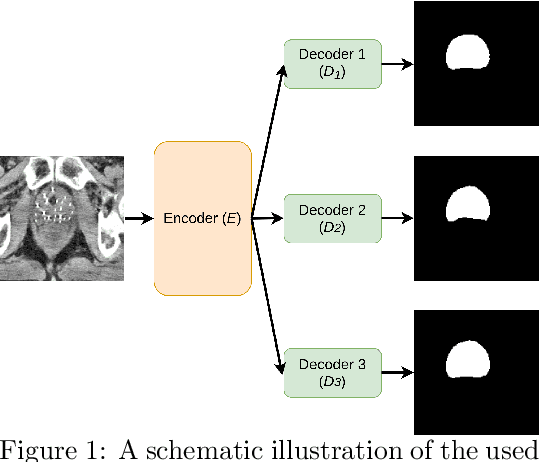
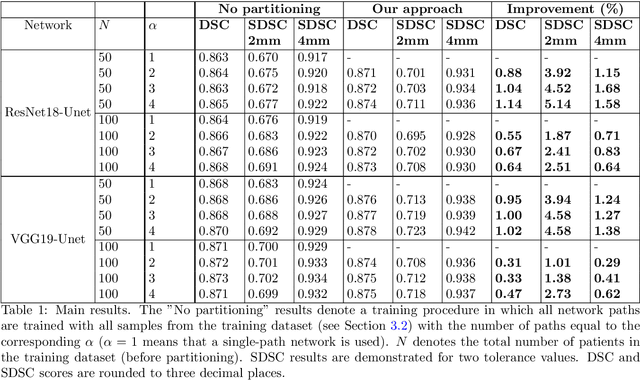
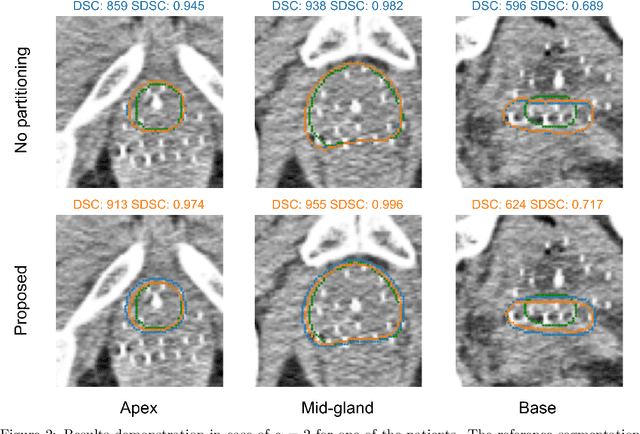
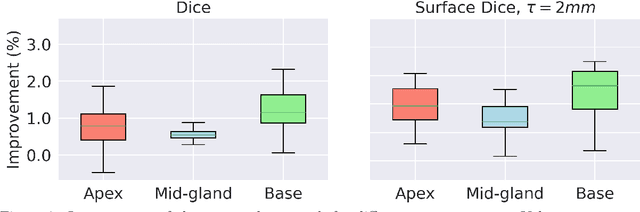
Abstract:Deep learning algorithms have become the golden standard for segmentation of medical imaging data. In most works, the variability and heterogeneity of real clinical data is acknowledged to still be a problem. One way to automatically overcome this is to capture and exploit this variation explicitly. Here, we propose an approach that improves on our previous work in this area and explain how it potentially can improve clinical acceptance of (semi-)automatic segmentation methods. In contrast to a standard neural network that produces one segmentation, we propose to use a multi-pathUnet network that produces multiple segmentation variants, presumably corresponding to the variations that reside in the dataset. Different paths of the network are trained on disjoint data subsets. Because a priori it may be unclear what variations exist in the data, the subsets should be automatically determined. This is achieved by searching for the best data partitioning with an evolutionary optimization algorithm. Because each network path can become more specialized when trained on a more homogeneous data subset, better segmentation quality can be achieved. In practical usage, various automatically produced segmentations can be presented to a medical expert, from which the preferred segmentation can be selected. In experiments with a real clinical dataset of CT scans with prostate segmentations, our approach provides an improvement of several percentage points in terms of Dice and surface Dice coefficients compared to when all network paths are trained on all training data. Noticeably, the largest improvement occurs in the upper part of the prostate that is known to be most prone to inter-observer segmentation variation.
Mixed-Block Neural Architecture Search for Medical Image Segmentation
Feb 23, 2022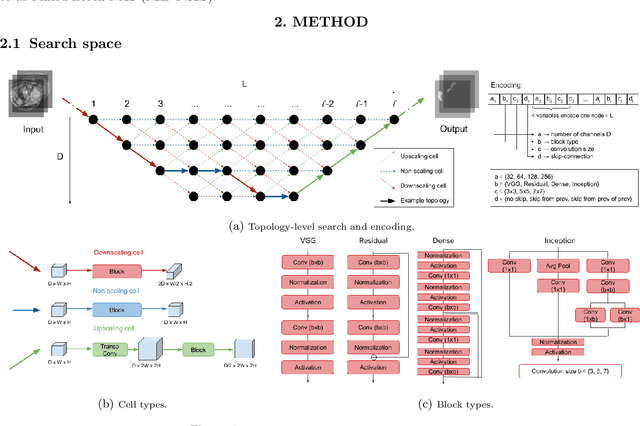
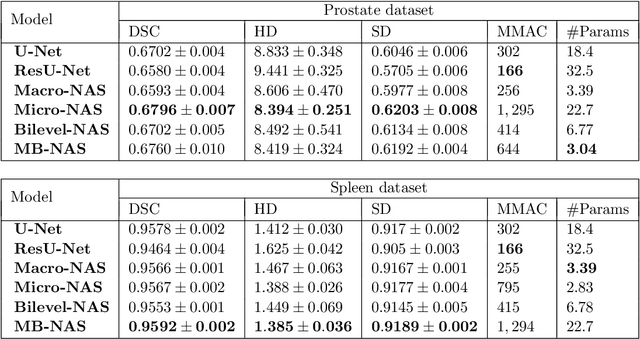
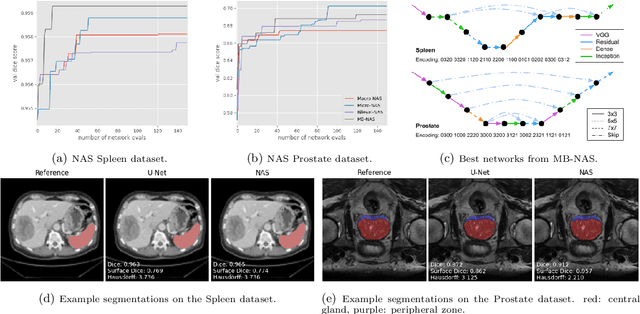
Abstract:Deep Neural Networks (DNNs) have the potential for making various clinical procedures more time-efficient by automating medical image segmentation. Due to their strong, in some cases human-level, performance, they have become the standard approach in this field. The design of the best possible medical image segmentation DNNs, however, is task-specific. Neural Architecture Search (NAS), i.e., the automation of neural network design, has been shown to have the capability to outperform manually designed networks for various tasks. However, the existing NAS methods for medical image segmentation have explored a quite limited range of types of DNN architectures that can be discovered. In this work, we propose a novel NAS search space for medical image segmentation networks. This search space combines the strength of a generalised encoder-decoder structure, well known from U-Net, with network blocks that have proven to have a strong performance in image classification tasks. The search is performed by looking for the best topology of multiple cells simultaneously with the configuration of each cell within, allowing for interactions between topology and cell-level attributes. From experiments on two publicly available datasets, we find that the networks discovered by our proposed NAS method have better performance than well-known handcrafted segmentation networks, and outperform networks found with other NAS approaches that perform only topology search, and topology-level search followed by cell-level search.
Heed the Noise in Performance Evaluations in Neural Architecture Search
Feb 04, 2022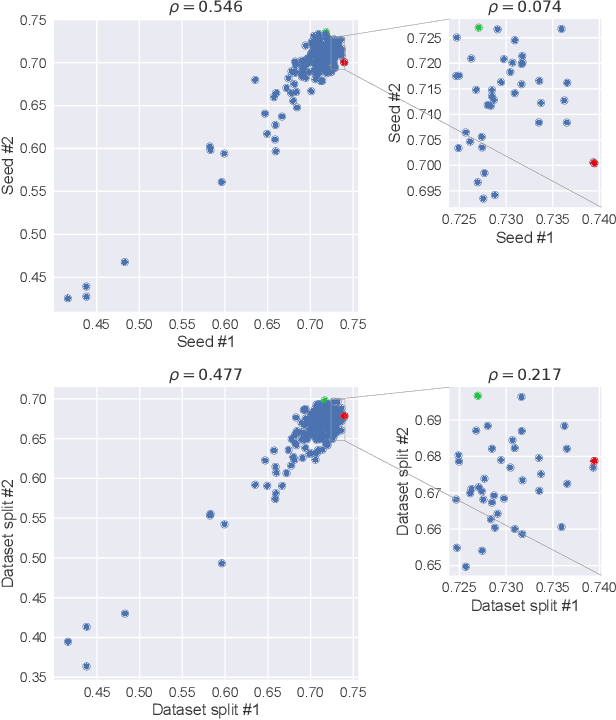
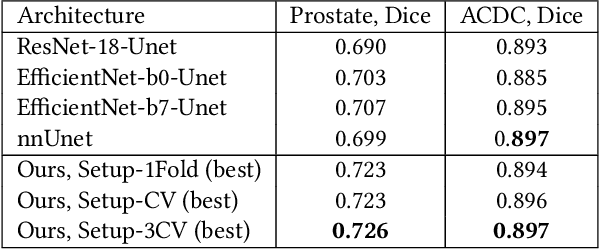
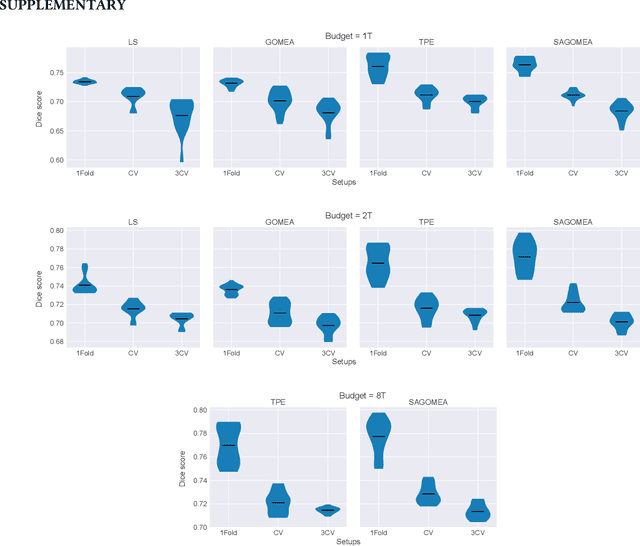
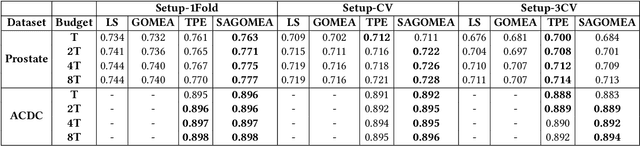
Abstract:Neural Architecture Search (NAS) has recently become a topic of great interest. However, there is a potentially impactful issue within NAS that remains largely unrecognized: noise. Due to stochastic factors in neural network initialization, training, and the chosen train/validation dataset split, the performance evaluation of a neural network architecture, which is often based on a single learning run, is also stochastic. This may have a particularly large impact if a dataset is small. We therefore propose to reduce the noise by having architecture evaluations comprise averaging of scores over multiple network training runs using different random seeds and cross-validation. We perform experiments for a combinatorial optimization formulation of NAS in which we vary noise reduction levels. We use the same computational budget for each noise level in terms of network training runs, i.e., we allow less architecture evaluations when averaging over more training runs. Multiple search algorithms are considered, including evolutionary algorithms which generally perform well for NAS. We use two publicly available datasets from the medical image segmentation domain where datasets are often limited and variability among samples is often high. Our results show that reducing noise in architecture evaluations enables finding better architectures by all considered search algorithms.
Parameterless Gene-pool Optimal Mixing Evolutionary Algorithms
Sep 11, 2021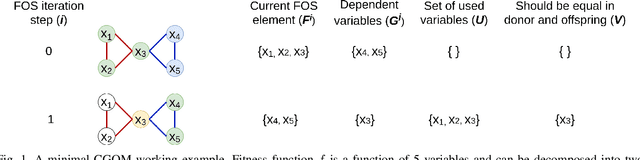
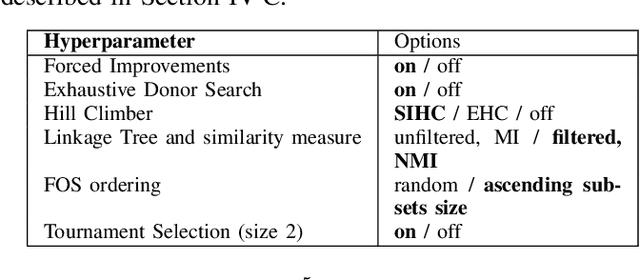
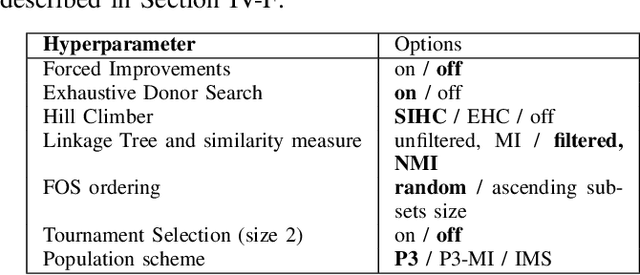
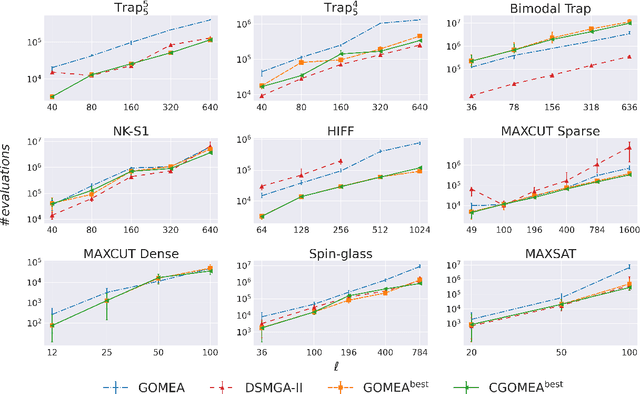
Abstract:When it comes to solving optimization problems with evolutionary algorithms (EAs) in a reliable and scalable manner, detecting and exploiting linkage information, i.e., dependencies between variables, can be key. In this article, we present the latest version of, and propose substantial enhancements to, the Gene-pool Optimal Mixing Evoutionary Algorithm (GOMEA): an EA explicitly designed to estimate and exploit linkage information. We begin by performing a large-scale search over several GOMEA design choices, to understand what matters most and obtain a generally best-performing version of the algorithm. Next, we introduce a novel version of GOMEA, called CGOMEA, where linkage-based variation is further improved by filtering solution mating based on conditional dependencies. We compare our latest version of GOMEA, the newly introduced CGOMEA, and another contending linkage-aware EA DSMGA-II in an extensive experimental evaluation, involving a benchmark set of 9 black-box problems that can only be solved efficiently if their inherent dependency structure is unveiled and exploited. Finally, in an attempt to make EAs more usable and resilient to parameter choices, we investigate the performance of different automatic population management schemes for GOMEA and CGOMEA, de facto making the EAs parameterless. Our results show that GOMEA and CGOMEA significantly outperform the original GOMEA and DSMGA-II on most problems, setting a new state of the art for the field.
A Novel Surrogate-assisted Evolutionary Algorithm Applied to Partition-based Ensemble Learning
Apr 16, 2021
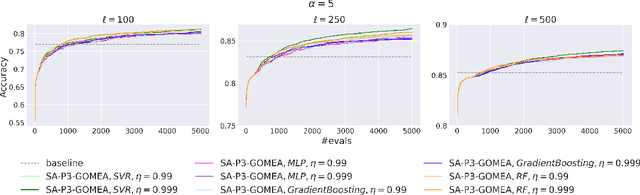
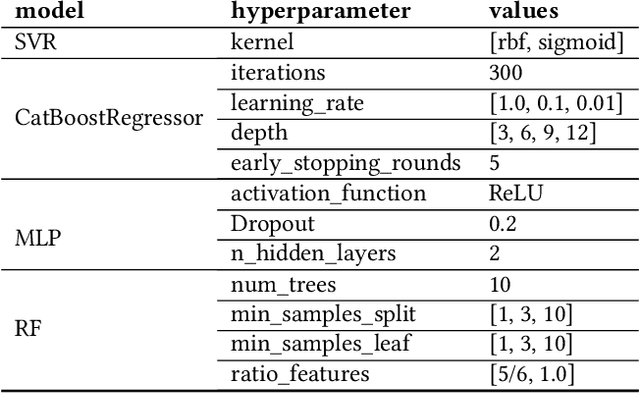
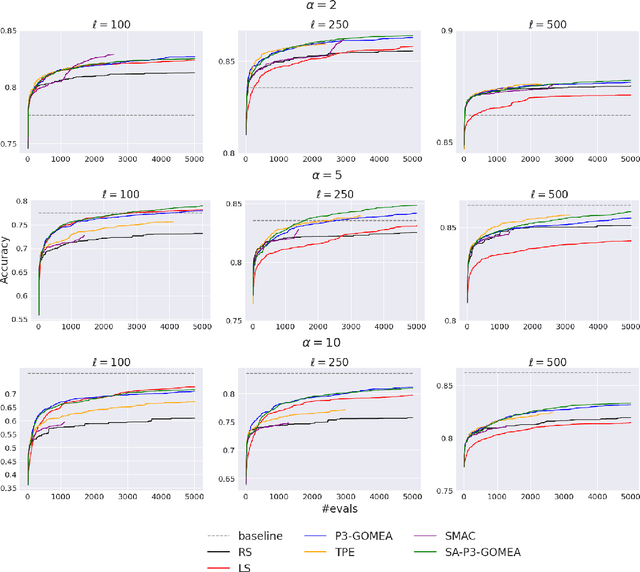
Abstract:We propose a novel surrogate-assisted Evolutionary Algorithm for solving expensive combinatorial optimization problems. We integrate a surrogate model, which is used for fitness value estimation, into a state-of-the-art P3-like variant of the Gene-Pool Optimal Mixing Algorithm (GOMEA) and adapt the resulting algorithm for solving non-binary combinatorial problems. We test the proposed algorithm on an ensemble learning problem. Ensembling several models is a common Machine Learning technique to achieve better performance. We consider ensembles of several models trained on disjoint subsets of a dataset. Finding the best dataset partitioning is naturally a combinatorial non-binary optimization problem. Fitness function evaluations can be extremely expensive if complex models, such as Deep Neural Networks, are used as learners in an ensemble. Therefore, the number of fitness function evaluations is typically limited, necessitating expensive optimization techniques. In our experiments we use five classification datasets from the OpenML-CC18 benchmark and Support-vector Machines as learners in an ensemble. The proposed algorithm demonstrates better performance than alternative approaches, including Bayesian optimization algorithms. It manages to find better solutions using just several thousand fitness function evaluations for an ensemble learning problem with up to 500 variables.
Observer variation-aware medical image segmentation by combining deep learning and surrogate-assisted genetic algorithms
Jan 23, 2020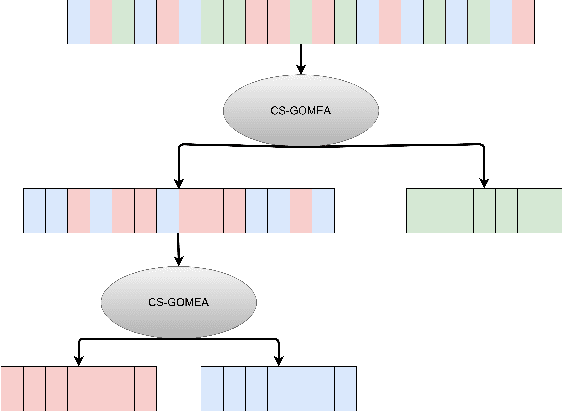
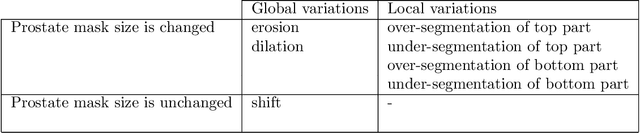
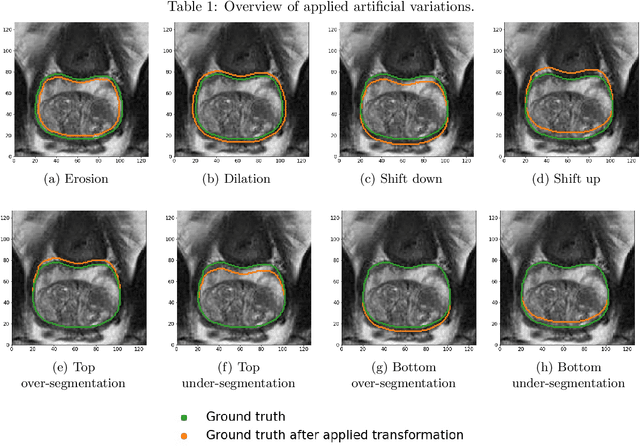
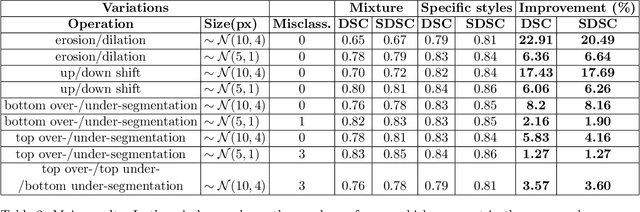
Abstract:There has recently been great progress in automatic segmentation of medical images with deep learning algorithms. In most works observer variation is acknowledged to be a problem as it makes training data heterogeneous but so far no attempts have been made to explicitly capture this variation. Here, we propose an approach capable of mimicking different styles of segmentation, which potentially can improve quality and clinical acceptance of automatic segmentation methods. In this work, instead of training one neural network on all available data, we train several neural networks on subgroups of data belonging to different segmentation variations separately. Because a priori it may be unclear what styles of segmentation exist in the data and because different styles do not necessarily map one-on-one to different observers, the subgroups should be automatically determined. We achieve this by searching for the best data partition with a genetic algorithm. Therefore, each network can learn a specific style of segmentation from grouped training data. We provide proof of principle results for open-sourced prostate segmentation MRI data with simulated observer variations. Our approach provides an improvement of up to 23% (depending on simulated variations) in terms of Dice and surface Dice coefficients compared to one network trained on all data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge