Dirk Thierens
Sharing Knowledge without Sharing Data: Stitches can improve ensembles of disjointly trained models
Dec 19, 2025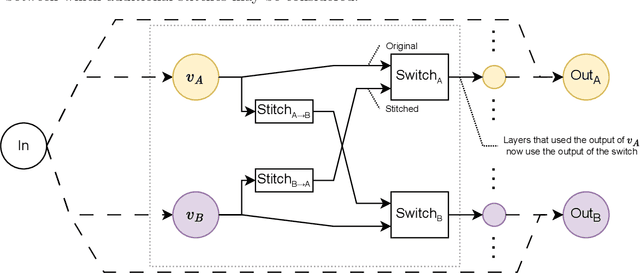
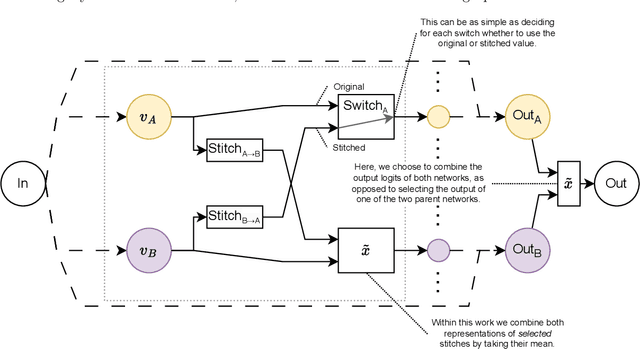
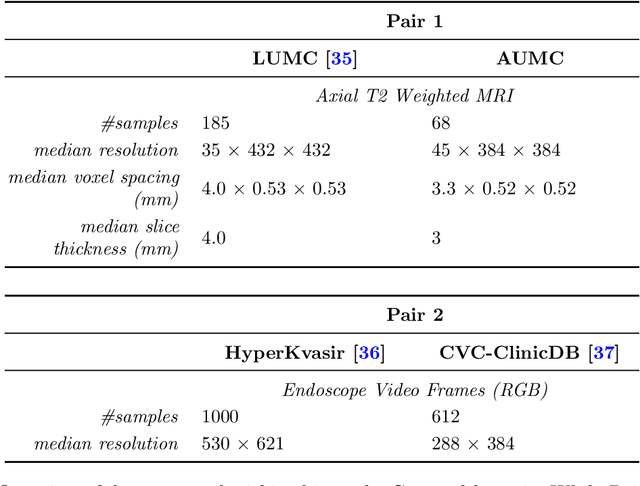
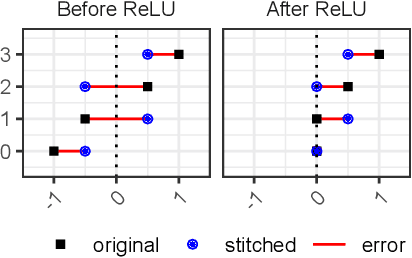
Abstract:Deep learning has been shown to be very capable at performing many real-world tasks. However, this performance is often dependent on the presence of large and varied datasets. In some settings, like in the medical domain, data is often fragmented across parties, and cannot be readily shared. While federated learning addresses this situation, it is a solution that requires synchronicity of parties training a single model together, exchanging information about model weights. We investigate how asynchronous collaboration, where only already trained models are shared (e.g. as part of a publication), affects performance, and propose to use stitching as a method for combining models. Through taking a multi-objective perspective, where performance on each parties' data is viewed independently, we find that training solely on a single parties' data results in similar performance when merging with another parties' data, when considering performance on that single parties' data, while performance on other parties' data is notably worse. Moreover, while an ensemble of such individually trained networks generalizes better, performance on each parties' own dataset suffers. We find that combining intermediate representations in individually trained models with a well placed pair of stitching layers allows this performance to recover to a competitive degree while maintaining improved generalization, showing that asynchronous collaboration can yield competitive results.
The Pitfalls and Potentials of Adding Gene-invariance to Optimal Mixing
Jun 18, 2025Abstract:Optimal Mixing (OM) is a variation operator that integrates local search with genetic recombination. EAs with OM are capable of state-of-the-art optimization in discrete spaces, offering significant advantages over classic recombination-based EAs. This success is partly due to high selection pressure that drives rapid convergence. However, this can also negatively impact population diversity, complicating the solving of hierarchical problems, which feature multiple layers of complexity. While there have been attempts to address this issue, these solutions are often complicated and prone to bias. To overcome this, we propose a solution inspired by the Gene Invariant Genetic Algorithm (GIGA), which preserves gene frequencies in the population throughout the process. This technique is tailored to and integrated with the Gene-pool Optimal Mixing Evolutionary Algorithm (GOMEA), resulting in GI-GOMEA. The simple, yet elegant changes are found to have striking potential: GI-GOMEA outperforms GOMEA on a range of well-known problems, even when these problems are adjusted for pitfalls - biases in much-used benchmark problems that can be easily exploited by maintaining gene invariance. Perhaps even more notably, GI-GOMEA is also found to be effective at solving hierarchical problems, including newly introduced asymmetric hierarchical trap functions.
Learning with Confidence: Training Better Classifiers from Soft Labels
Sep 24, 2024



Abstract:In supervised machine learning, models are typically trained using data with hard labels, i.e., definite assignments of class membership. This traditional approach, however, does not take the inherent uncertainty in these labels into account. We investigate whether incorporating label uncertainty, represented as discrete probability distributions over the class labels -- known as soft labels -- improves the predictive performance of classification models. We first demonstrate the potential value of soft label learning (SLL) for estimating model parameters in a simulation experiment, particularly for limited sample sizes and imbalanced data. Subsequently, we compare the performance of various wrapper methods for learning from both hard and soft labels using identical base classifiers. On real-world-inspired synthetic data with clean labels, the SLL methods consistently outperform hard label methods. Since real-world data is often noisy and precise soft labels are challenging to obtain, we study the effect that noisy probability estimates have on model performance. Alongside conventional noise models, our study examines four types of miscalibration that are known to affect human annotators. The results show that SLL methods outperform the hard label methods in the majority of settings. Finally, we evaluate the methods on a real-world dataset with confidence scores, where the SLL methods are shown to match the traditional methods for predicting the (noisy) hard labels while providing more accurate confidence estimates.
Stitching for Neuroevolution: Recombining Deep Neural Networks without Breaking Them
Mar 21, 2024


Abstract:Traditional approaches to neuroevolution often start from scratch. This becomes prohibitively expensive in terms of computational and data requirements when targeting modern, deep neural networks. Using a warm start could be highly advantageous, e.g., using previously trained networks, potentially from different sources. This moreover enables leveraging the benefits of transfer learning (in particular vastly reduced training effort). However, recombining trained networks is non-trivial because architectures and feature representations typically differ. Consequently, a straightforward exchange of layers tends to lead to a performance breakdown. We overcome this by matching the layers of parent networks based on their connectivity, identifying potential crossover points. To correct for differing feature representations between these layers we employ stitching, which merges the networks by introducing new layers at crossover points. To train the merged network, only stitching layers need to be considered. New networks can then be created by selecting a subnetwork by choosing which stitching layers to (not) use. Assessing their performance is efficient as only their evaluation on data is required. We experimentally show that our approach enables finding networks that represent novel trade-offs between performance and computational cost, with some even dominating the original networks.
Generating the Ground Truth: Synthetic Data for Label Noise Research
Sep 08, 2023Abstract:Most real-world classification tasks suffer from label noise to some extent. Such noise in the data adversely affects the generalization error of learned models and complicates the evaluation of noise-handling methods, as their performance cannot be accurately measured without clean labels. In label noise research, typically either noisy or incomplex simulated data are accepted as a baseline, into which additional noise with known properties is injected. In this paper, we propose SYNLABEL, a framework that aims to improve upon the aforementioned methodologies. It allows for creating a noiseless dataset informed by real data, by either pre-specifying or learning a function and defining it as the ground truth function from which labels are generated. Furthermore, by resampling a number of values for selected features in the function domain, evaluating the function and aggregating the resulting labels, each data point can be assigned a soft label or label distribution. Such distributions allow for direct injection and quantification of label noise. The generated datasets serve as a clean baseline of adjustable complexity into which different types of noise may be introduced. We illustrate how the framework can be applied, how it enables quantification of label noise and how it improves over existing methodologies.
The Impact of Asynchrony on Parallel Model-Based EAs
Mar 27, 2023



Abstract:In a parallel EA one can strictly adhere to the generational clock, and wait for all evaluations in a generation to be done. However, this idle time limits the throughput of the algorithm and wastes computational resources. Alternatively, an EA can be made asynchronous parallel. However, EAs using classic recombination and selection operators (GAs) are known to suffer from an evaluation time bias, which also influences the performance of the approach. Model-Based Evolutionary Algorithms (MBEAs) are more scalable than classic GAs by virtue of capturing the structure of a problem in a model. If this model is learned through linkage learning based on the population, the learned model may also capture biases. Thus, if an asynchronous parallel MBEA is also affected by an evaluation time bias, this could result in learned models to be less suited to solving the problem, reducing performance. Therefore, in this work, we study the impact and presence of evaluation time biases on MBEAs in an asynchronous parallelization setting, and compare this to the biases in GAs. We find that a modern MBEA, GOMEA, is unaffected by evaluation time biases, while the more classical MBEA, ECGA, is affected, much like GAs are.
Solving Multi-Structured Problems by Introducing Linkage Kernels into GOMEA
Mar 11, 2022



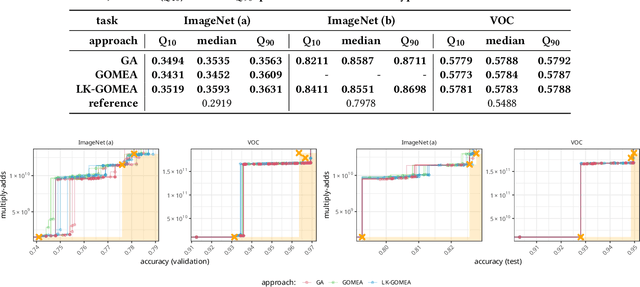
Abstract:Model-Based Evolutionary Algorithms (MBEAs) can be highly scalable by virtue of linkage (or variable interaction) learning. This requires, however, that the linkage model can capture the exploitable structure of a problem. Usually, a single type of linkage structure is attempted to be captured using models such as a linkage tree. However, in practice, problems may exhibit multiple linkage structures. This is for instance the case in multi-objective optimization when the objectives have different linkage structures. This cannot be modelled sufficiently well when using linkage models that aim at capturing a single type of linkage structure, deteriorating the advantages brought by MBEAs. Therefore, here, we introduce linkage kernels, whereby a linkage structure is learned for each solution over its local neighborhood. We implement linkage kernels into the MBEA known as GOMEA that was previously found to be highly scalable when solving various problems. We further introduce a novel benchmark function called Best-of-Traps (BoT) that has an adjustable degree of different linkage structures. On both BoT and a worst-case scenario-based variant of the well-known MaxCut problem, we experimentally find a vast performance improvement of linkage-kernel GOMEA over GOMEA with a single linkage tree as well as the MBEA known as DSMGA-II.
Parameterless Gene-pool Optimal Mixing Evolutionary Algorithms
Sep 11, 2021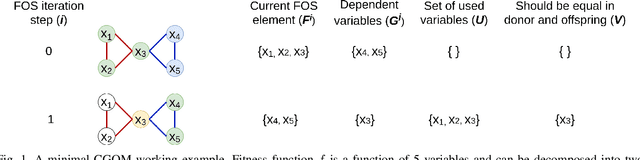
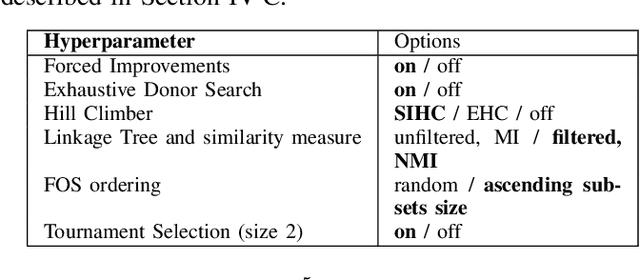
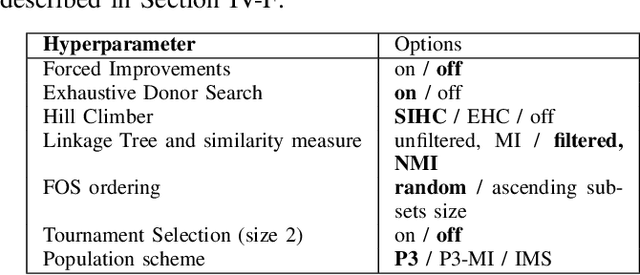
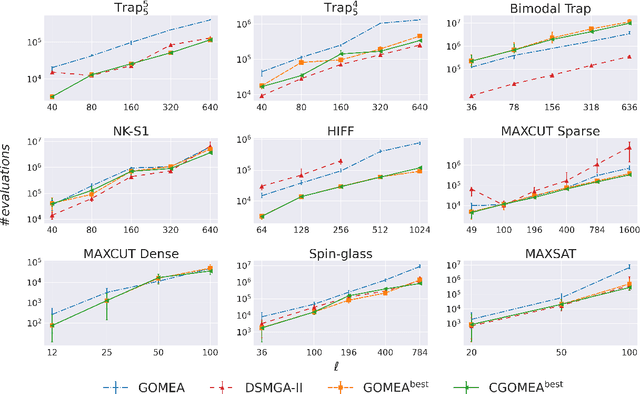
Abstract:When it comes to solving optimization problems with evolutionary algorithms (EAs) in a reliable and scalable manner, detecting and exploiting linkage information, i.e., dependencies between variables, can be key. In this article, we present the latest version of, and propose substantial enhancements to, the Gene-pool Optimal Mixing Evoutionary Algorithm (GOMEA): an EA explicitly designed to estimate and exploit linkage information. We begin by performing a large-scale search over several GOMEA design choices, to understand what matters most and obtain a generally best-performing version of the algorithm. Next, we introduce a novel version of GOMEA, called CGOMEA, where linkage-based variation is further improved by filtering solution mating based on conditional dependencies. We compare our latest version of GOMEA, the newly introduced CGOMEA, and another contending linkage-aware EA DSMGA-II in an extensive experimental evaluation, involving a benchmark set of 9 black-box problems that can only be solved efficiently if their inherent dependency structure is unveiled and exploited. Finally, in an attempt to make EAs more usable and resilient to parameter choices, we investigate the performance of different automatic population management schemes for GOMEA and CGOMEA, de facto making the EAs parameterless. Our results show that GOMEA and CGOMEA significantly outperform the original GOMEA and DSMGA-II on most problems, setting a new state of the art for the field.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge