Anton Bouter
The Pitfalls and Potentials of Adding Gene-invariance to Optimal Mixing
Jun 18, 2025Abstract:Optimal Mixing (OM) is a variation operator that integrates local search with genetic recombination. EAs with OM are capable of state-of-the-art optimization in discrete spaces, offering significant advantages over classic recombination-based EAs. This success is partly due to high selection pressure that drives rapid convergence. However, this can also negatively impact population diversity, complicating the solving of hierarchical problems, which feature multiple layers of complexity. While there have been attempts to address this issue, these solutions are often complicated and prone to bias. To overcome this, we propose a solution inspired by the Gene Invariant Genetic Algorithm (GIGA), which preserves gene frequencies in the population throughout the process. This technique is tailored to and integrated with the Gene-pool Optimal Mixing Evolutionary Algorithm (GOMEA), resulting in GI-GOMEA. The simple, yet elegant changes are found to have striking potential: GI-GOMEA outperforms GOMEA on a range of well-known problems, even when these problems are adjusted for pitfalls - biases in much-used benchmark problems that can be easily exploited by maintaining gene invariance. Perhaps even more notably, GI-GOMEA is also found to be effective at solving hierarchical problems, including newly introduced asymmetric hierarchical trap functions.
A Tournament of Transformation Models: B-Spline-based vs. Mesh-based Multi-Objective Deformable Image Registration
Jan 30, 2024Abstract:The transformation model is an essential component of any deformable image registration approach. It provides a representation of physical deformations between images, thereby defining the range and realism of registrations that can be found. Two types of transformation models have emerged as popular choices: B-spline models and mesh models. Although both models have been investigated in detail, a direct comparison has not yet been made, since the models are optimized using very different optimization methods in practice. B-spline models are predominantly optimized using gradient-descent methods, while mesh models are typically optimized using finite-element method solvers or evolutionary algorithms. Multi-objective optimization methods, which aim to find a diverse set of high-quality trade-off registrations, are increasingly acknowledged to be important in deformable image registration. Since these methods search for a diverse set of registrations, they can provide a more complete picture of the capabilities of different transformation models, making them suitable for a comparison of models. In this work, we conduct the first direct comparison between B-spline and mesh transformation models, by optimizing both models with the same state-of-the-art multi-objective optimization method, the Multi-Objective Real-Valued Gene-pool Optimal Mixing Evolutionary Algorithm (MO-RV-GOMEA). The combination with B-spline transformation models, moreover, is novel. We experimentally compare both models on two different registration problems that are both based on pelvic CT scans of cervical cancer patients, featuring large deformations. Our results, on three cervical cancer patients, indicate that the choice of transformation model can have a profound impact on the diversity and quality of achieved registration outcomes.
A Joint Python/C++ Library for Efficient yet Accessible Black-Box and Gray-Box Optimization with GOMEA
May 10, 2023Abstract:Exploiting knowledge about the structure of a problem can greatly benefit the efficiency and scalability of an Evolutionary Algorithm (EA). Model-Based EAs (MBEAs) are capable of doing this by explicitly modeling the problem structure. The Gene-pool Optimal Mixing Evolutionary Algorithm (GOMEA) is among the state-of-the-art of MBEAs due to its use of a linkage model and the optimal mixing variation operator. Especially in a Gray-Box Optimization (GBO) setting that allows for partial evaluations, i.e., the relatively efficient evaluation of a partial modification of a solution, GOMEA is known to excel. Such GBO settings are known to exist in various real-world applications to which GOMEA has successfully been applied. In this work, we introduce the GOMEA library, making existing GOMEA code in C++ accessible through Python, which serves as a centralized way of maintaining and distributing code of GOMEA for various optimization domains. Moreover, it allows for the straightforward definition of BBO as well as GBO fitness functions within Python, which are called from the C++ optimization code for each required (partial) evaluation. We describe the structure of the GOMEA library and how it can be used, and we show its performance in both GBO and Black-Box Optimization (BBO).
Obtaining Smoothly Navigable Approximation Sets in Bi-Objective Multi-Modal Optimization
Mar 24, 2022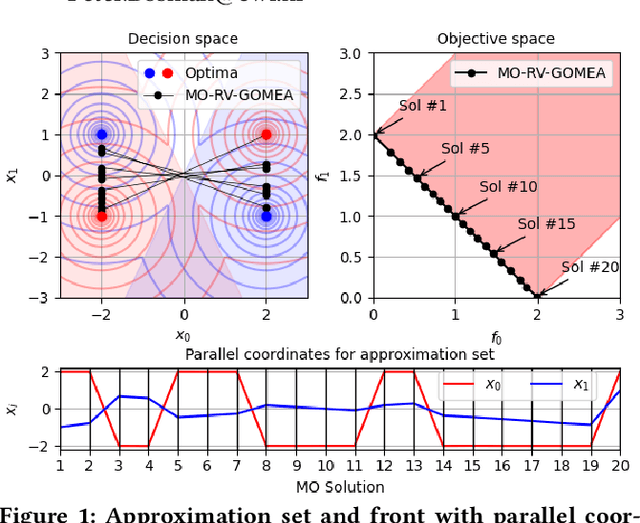
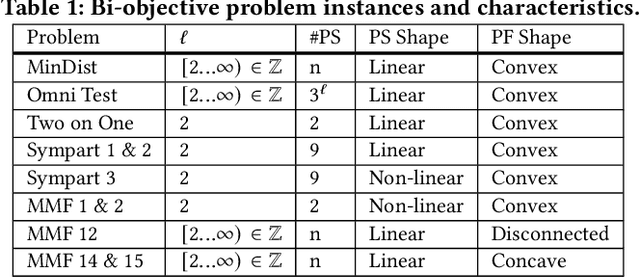
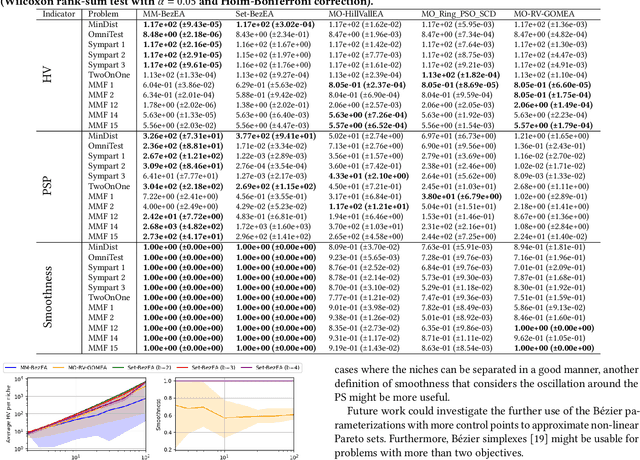
Abstract:Even if a Multi-modal Multi-Objective Evolutionary Algorithm (MMOEA) is designed to find all locally optimal approximation sets of a Multi-modal Multi-objective Optimization Problem (MMOP), there is a risk that the found approximation sets are not smoothly navigable because the solutions belong to various niches, reducing the insight for decision makers. Moreover, when the multi-modality of MMOPs increases, this risk grows and the trackability of finding all locally optimal approximation sets decreases. To tackle these issues, two new MMOEAs are proposed: Multi-Modal B\'ezier Evolutionary Algorithm (MM-BezEA) and Set B\'ezier Evolutionary Algorithm (Set-BezEA). Both MMOEAs produce approximation sets that cover individual niches and exhibit inherent decision-space smoothness as they are parameterized by B\'ezier curves. MM-BezEA combines the concepts behind the recently introduced BezEA and MO-HillVallEA to find all locally optimal approximation sets. Set-BezEA employs a novel multi-objective fitness function formulation to find limited numbers of diverse, locally optimal, approximation sets for MMOPs of high multi-modality. Both algorithms, but especially MM-BezEA, are found to outperform the MMOEAs MO_Ring_PSO_SCD and MO-HillVallEA on MMOPs of moderate multi-modality with linear Pareto sets. Moreover, for MMOPs of high multi-modality, Set-BezEA is found to indeed be able to produce high-quality approximation sets, each pertaining to a single niche.
GPU-Accelerated Parallel Gene-pool Optimal Mixing in a Gray-Box Optimization Setting
Mar 16, 2022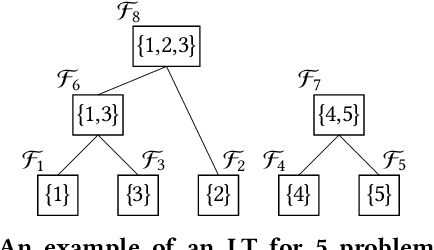
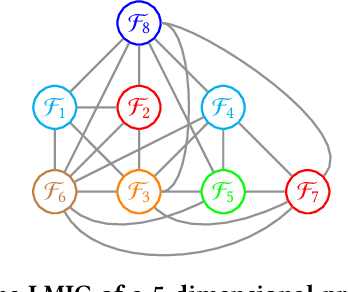
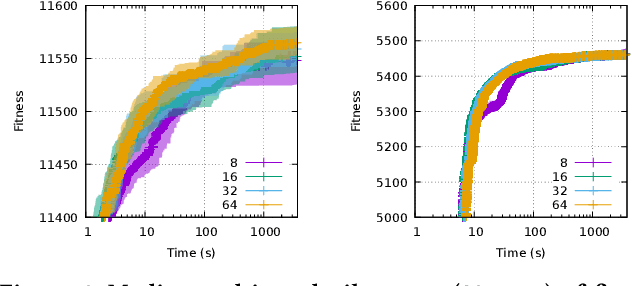

Abstract:In a Gray-Box Optimization (GBO) setting that allows for partial evaluations, the fitness of an individual can be updated efficiently after a subset of its variables has been modified. This enables more efficient evolutionary optimization with the Gene-pool Optimal Mixing Evolutionary Algorithm (GOMEA) due to its key strength: Gene-pool Optimal Mixing (GOM). For each solution, GOM performs variation for many (small) sets of variables. To improve efficiency even further, parallel computing can be leveraged. For EAs, typically, this comprises population-wise parallelization. However, unless population sizes are large, this offers limited gains. For large GBO problems, parallelizing GOM-based variation holds greater speed-up potential, regardless of population size. However, this potential cannot be directly exploited because of dependencies between variables. We show how graph coloring can be used to group sets of variables that can undergo variation in parallel without violating dependencies. We test the performance of a CUDA implementation of parallel GOM on a Graphics Processing Unit (GPU) for the Max-Cut problem, a well-known problem for which the dependency structure can be controlled. We find that, for sufficiently large graphs with limited connectivity, finding high-quality solutions can be achieved up to 100 times faster, showcasing the great potential of our approach.
Parameterless Gene-pool Optimal Mixing Evolutionary Algorithms
Sep 11, 2021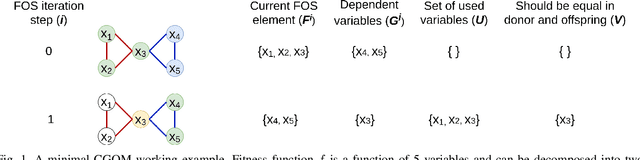
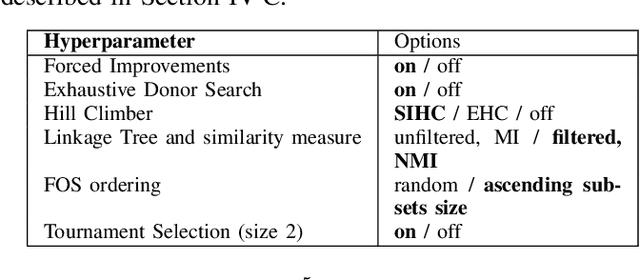
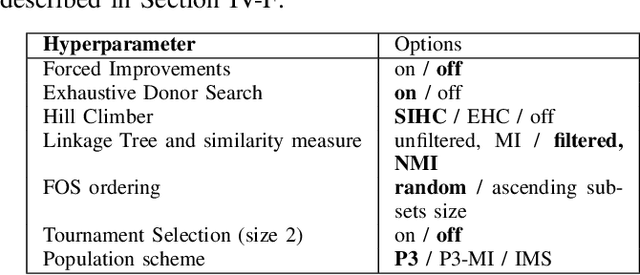
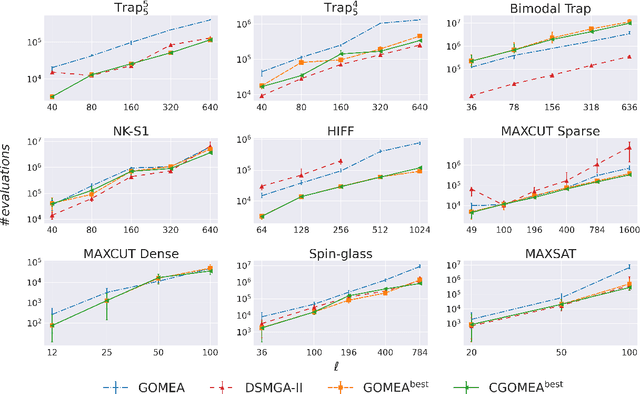
Abstract:When it comes to solving optimization problems with evolutionary algorithms (EAs) in a reliable and scalable manner, detecting and exploiting linkage information, i.e., dependencies between variables, can be key. In this article, we present the latest version of, and propose substantial enhancements to, the Gene-pool Optimal Mixing Evoutionary Algorithm (GOMEA): an EA explicitly designed to estimate and exploit linkage information. We begin by performing a large-scale search over several GOMEA design choices, to understand what matters most and obtain a generally best-performing version of the algorithm. Next, we introduce a novel version of GOMEA, called CGOMEA, where linkage-based variation is further improved by filtering solution mating based on conditional dependencies. We compare our latest version of GOMEA, the newly introduced CGOMEA, and another contending linkage-aware EA DSMGA-II in an extensive experimental evaluation, involving a benchmark set of 9 black-box problems that can only be solved efficiently if their inherent dependency structure is unveiled and exploited. Finally, in an attempt to make EAs more usable and resilient to parameter choices, we investigate the performance of different automatic population management schemes for GOMEA and CGOMEA, de facto making the EAs parameterless. Our results show that GOMEA and CGOMEA significantly outperform the original GOMEA and DSMGA-II on most problems, setting a new state of the art for the field.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge