Adriënne M. Mendrik
The Data Representativeness Criterion: Predicting the Performance of Supervised Classification Based on Data Set Similarity
Feb 27, 2020
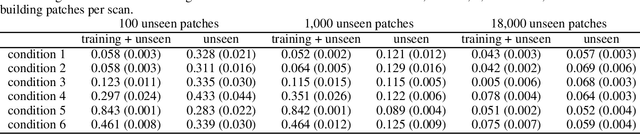
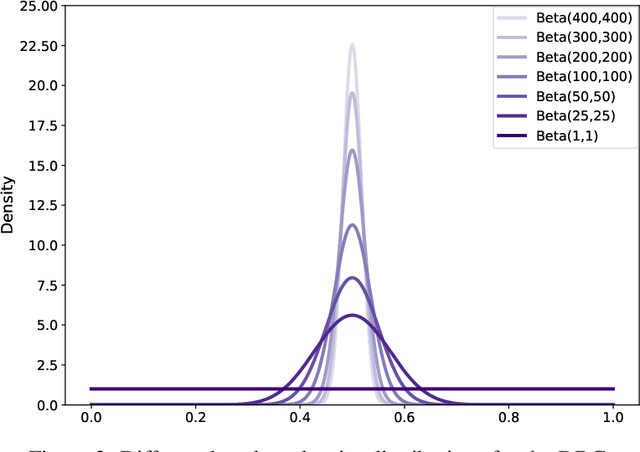
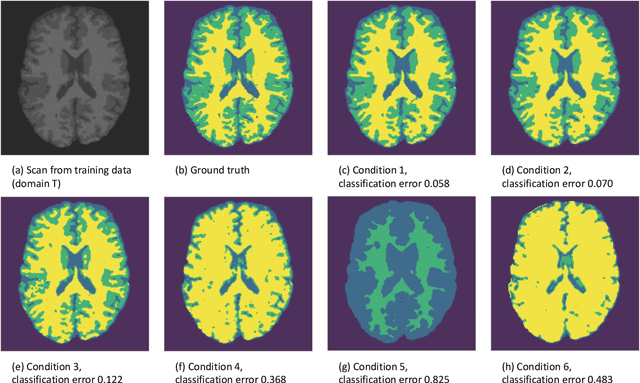
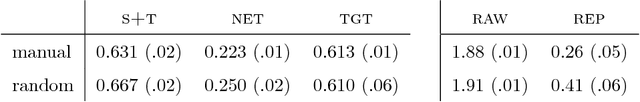
Abstract:In a broad range of fields it may be desirable to reuse a supervised classification algorithm and apply it to a new data set. However, generalization of such an algorithm and thus achieving a similar classification performance is only possible when the training data used to build the algorithm is similar to new unseen data one wishes to apply it to. It is often unknown in advance how an algorithm will perform on new unseen data, being a crucial reason for not deploying an algorithm at all. Therefore, tools are needed to measure the similarity of data sets. In this paper, we propose the Data Representativeness Criterion (DRC) to determine how representative a training data set is of a new unseen data set. We present a proof of principle, to see whether the DRC can quantify the similarity of data sets and whether the DRC relates to the performance of a supervised classification algorithm. We compared a number of magnetic resonance imaging (MRI) data sets, ranging from subtle to severe difference is acquisition parameters. Results indicate that, based on the similarity of data sets, the DRC is able to give an indication as to when the performance of a supervised classifier decreases. The strictness of the DRC can be set by the user, depending on what one considers to be an acceptable underperformance.
Observer variation-aware medical image segmentation by combining deep learning and surrogate-assisted genetic algorithms
Jan 23, 2020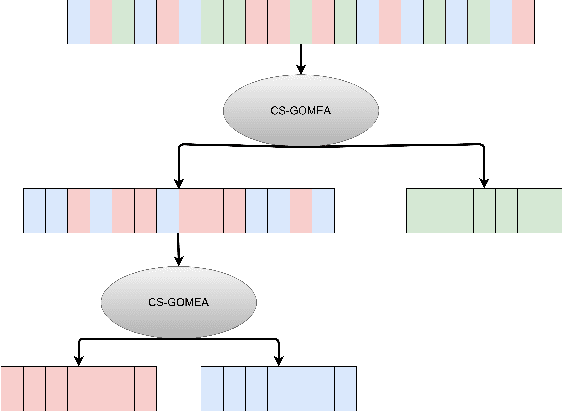
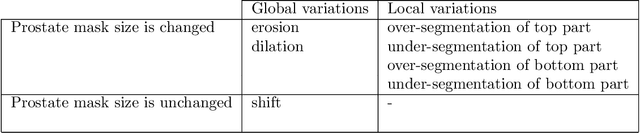
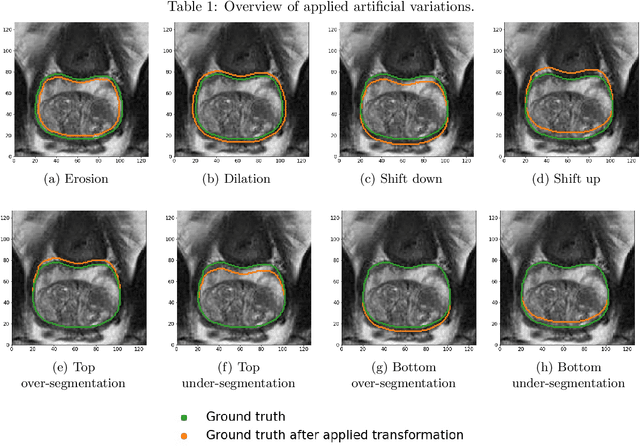
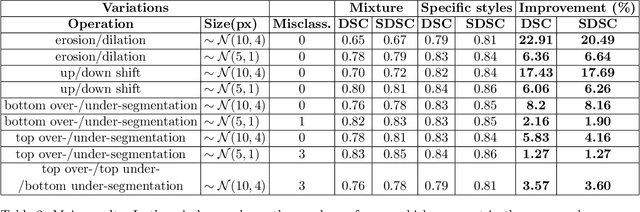
Abstract:There has recently been great progress in automatic segmentation of medical images with deep learning algorithms. In most works observer variation is acknowledged to be a problem as it makes training data heterogeneous but so far no attempts have been made to explicitly capture this variation. Here, we propose an approach capable of mimicking different styles of segmentation, which potentially can improve quality and clinical acceptance of automatic segmentation methods. In this work, instead of training one neural network on all available data, we train several neural networks on subgroups of data belonging to different segmentation variations separately. Because a priori it may be unclear what styles of segmentation exist in the data and because different styles do not necessarily map one-on-one to different observers, the subgroups should be automatically determined. We achieve this by searching for the best data partition with a genetic algorithm. Therefore, each network can learn a specific style of segmentation from grouped training data. We provide proof of principle results for open-sourced prostate segmentation MRI data with simulated observer variations. Our approach provides an improvement of up to 23% (depending on simulated variations) in terms of Dice and surface Dice coefficients compared to one network trained on all data.
Learning an MR acquisition-invariant representation using Siamese neural networks
Oct 17, 2018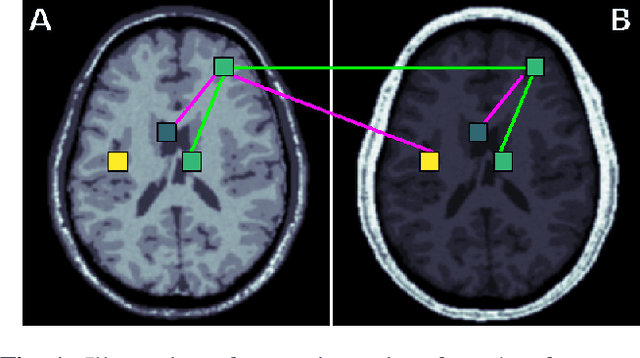
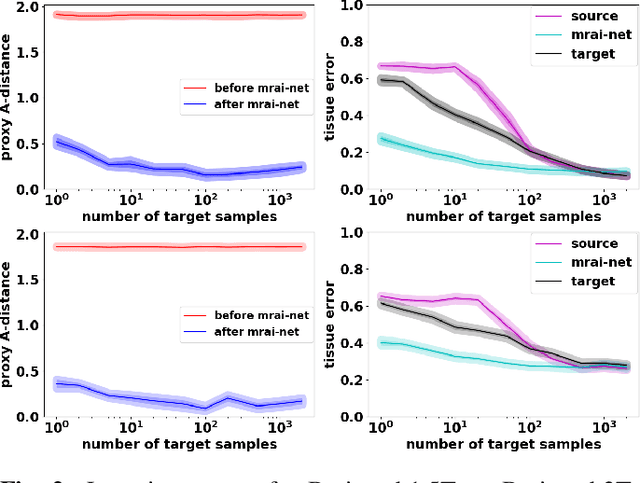
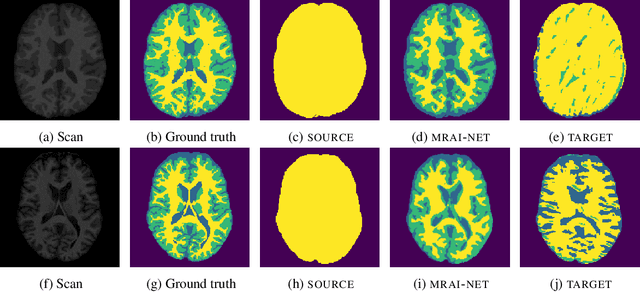
Abstract:Generalization of voxelwise classifiers is hampered by differences between MRI-scanners, e.g. different acquisition protocols and field strengths. To address this limitation, we propose a Siamese neural network (MRAI-NET) that extracts acquisition-invariant feature vectors. These can consequently be used by task-specific methods, such as voxelwise classifiers for tissue segmentation. MRAI-NET is tested on both simulated and real patient data. Experiments show that MRAI-NET outperforms voxelwise classifiers trained on the source or target scanner data when a small number of labeled samples is available.
MR Acquisition-Invariant Representation Learning
Apr 19, 2018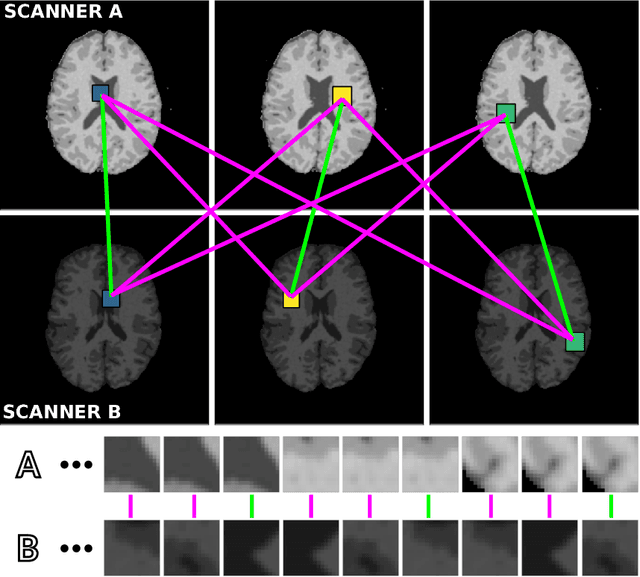

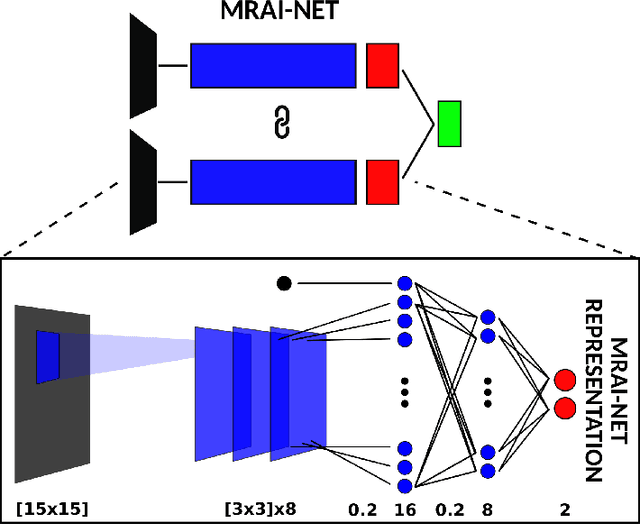
Abstract:Voxelwise classification approaches are popular and effective methods for tissue quantification in brain magnetic resonance imaging (MRI) scans. However, generalization of these approaches is hampered by large differences between sets of MRI scans such as differences in field strength, vendor or acquisition protocols. Due to this acquisition related variation, classifiers trained on data from a specific scanner fail or under-perform when applied to data that was acquired differently. In order to address this lack of generalization, we propose a Siamese neural network (MRAI-net) to learn a representation that minimizes the between-scanner variation, while maintaining the contrast between brain tissues necessary for brain tissue quantification. The proposed MRAI-net was evaluated on both simulated and real MRI data. After learning the MR acquisition invariant representation, any supervised classification model that uses feature vectors can be applied. In this paper, we provide a proof of principle, which shows that a linear classifier applied on the MRAI representation is able to outperform supervised convolutional neural network classifiers for tissue classification when little target training data is available.
Automatic segmentation of MR brain images with a convolutional neural network
Apr 11, 2017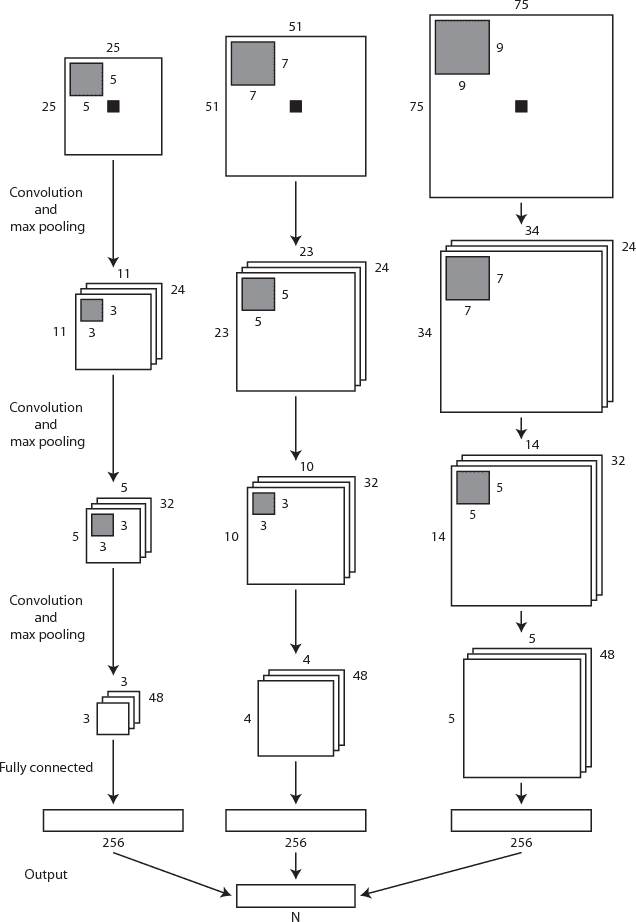

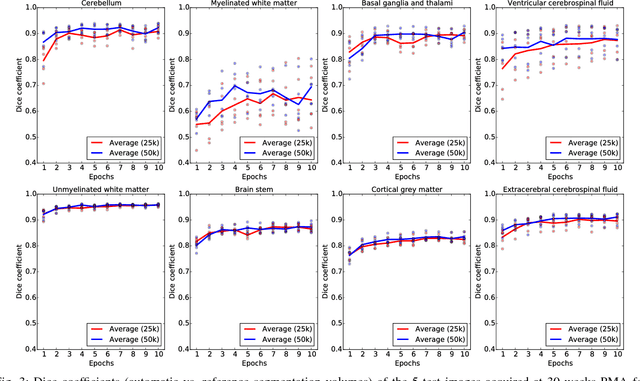

Abstract:Automatic segmentation in MR brain images is important for quantitative analysis in large-scale studies with images acquired at all ages. This paper presents a method for the automatic segmentation of MR brain images into a number of tissue classes using a convolutional neural network. To ensure that the method obtains accurate segmentation details as well as spatial consistency, the network uses multiple patch sizes and multiple convolution kernel sizes to acquire multi-scale information about each voxel. The method is not dependent on explicit features, but learns to recognise the information that is important for the classification based on training data. The method requires a single anatomical MR image only. The segmentation method is applied to five different data sets: coronal T2-weighted images of preterm infants acquired at 30 weeks postmenstrual age (PMA) and 40 weeks PMA, axial T2- weighted images of preterm infants acquired at 40 weeks PMA, axial T1-weighted images of ageing adults acquired at an average age of 70 years, and T1-weighted images of young adults acquired at an average age of 23 years. The method obtained the following average Dice coefficients over all segmented tissue classes for each data set, respectively: 0.87, 0.82, 0.84, 0.86 and 0.91. The results demonstrate that the method obtains accurate segmentations in all five sets, and hence demonstrates its robustness to differences in age and acquisition protocol.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge