Lambertus W. Bartels
MR Acquisition-Invariant Representation Learning
Apr 19, 2018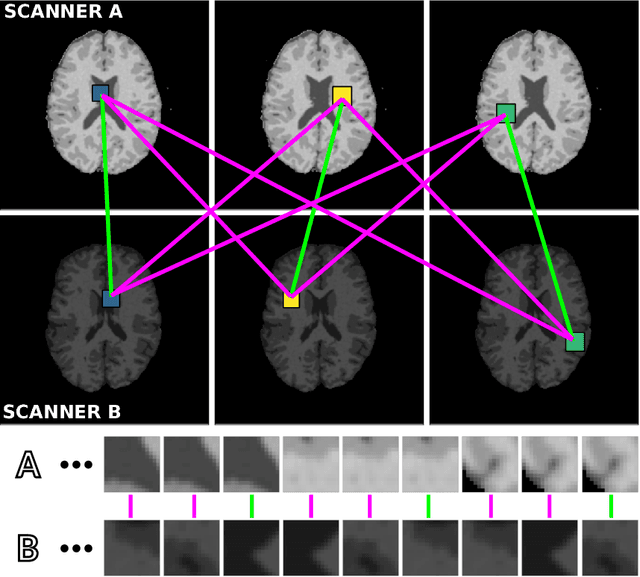

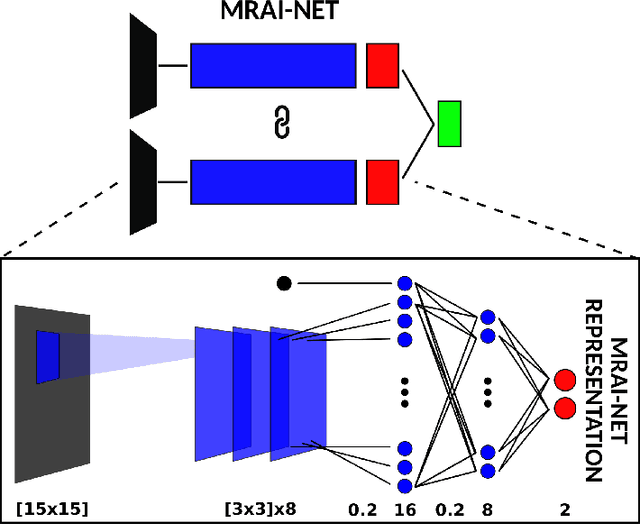
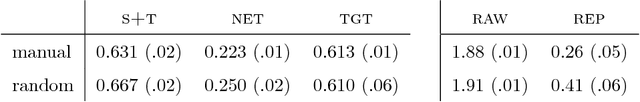
Abstract:Voxelwise classification approaches are popular and effective methods for tissue quantification in brain magnetic resonance imaging (MRI) scans. However, generalization of these approaches is hampered by large differences between sets of MRI scans such as differences in field strength, vendor or acquisition protocols. Due to this acquisition related variation, classifiers trained on data from a specific scanner fail or under-perform when applied to data that was acquired differently. In order to address this lack of generalization, we propose a Siamese neural network (MRAI-net) to learn a representation that minimizes the between-scanner variation, while maintaining the contrast between brain tissues necessary for brain tissue quantification. The proposed MRAI-net was evaluated on both simulated and real MRI data. After learning the MR acquisition invariant representation, any supervised classification model that uses feature vectors can be applied. In this paper, we provide a proof of principle, which shows that a linear classifier applied on the MRAI representation is able to outperform supervised convolutional neural network classifiers for tissue classification when little target training data is available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge