Timo M. Deist
Multi-Objective Learning to Predict Pareto Fronts Using Hypervolume Maximization
Feb 08, 2021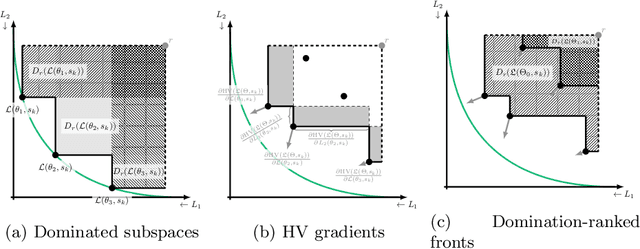
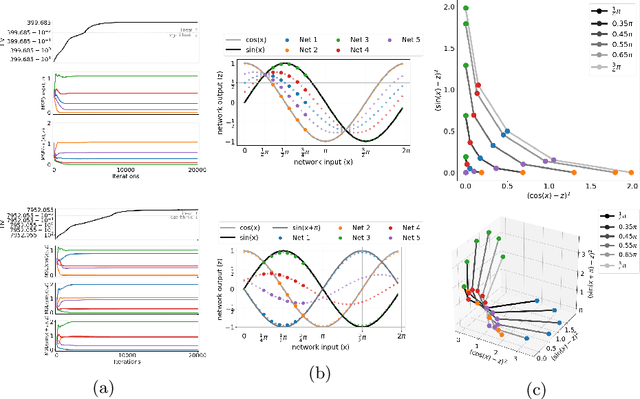
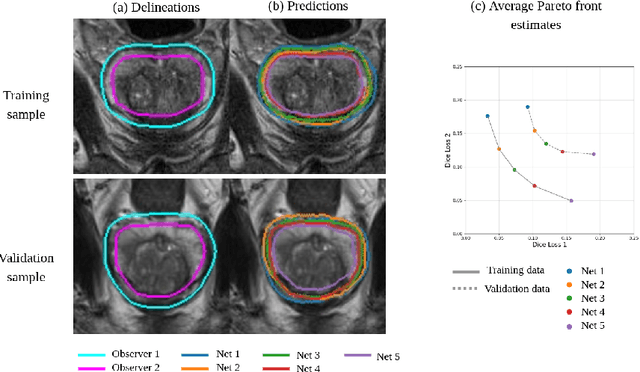
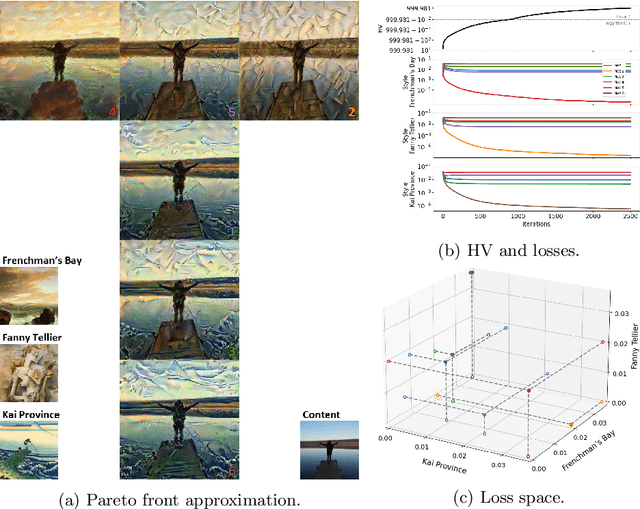
Abstract:Real-world problems are often multi-objective with decision-makers unable to specify a priori which trade-off between the conflicting objectives is preferable. Intuitively, building machine learning solutions in such cases would entail providing multiple predictions that span and uniformly cover the Pareto front of all optimal trade-off solutions. We propose a novel learning approach to estimate the Pareto front by maximizing the dominated hypervolume (HV) of the average loss vectors corresponding to a set of learners, leveraging established multi-objective optimization methods. In our approach, the set of learners are trained multi-objectively with a dynamic loss function, wherein each learner's losses are weighted by their HV maximizing gradients. Consequently, the learners get trained according to different trade-offs on the Pareto front, which otherwise is not guaranteed for fixed linear scalarizations or when optimizing for specific trade-offs per learner without knowing the shape of the Pareto front. Experiments on three different multi-objective tasks show that the outputs of the set of learners are indeed well-spread on the Pareto front. Further, the outputs corresponding to validation samples are also found to closely follow the trade-offs that were learned from training samples for our set of benchmark problems.
Identifying Properties of Real-World Optimisation Problems through a Questionnaire
Nov 11, 2020
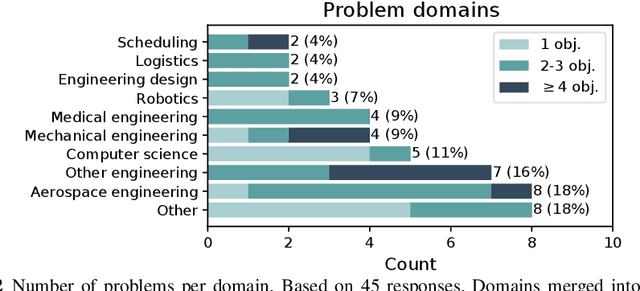
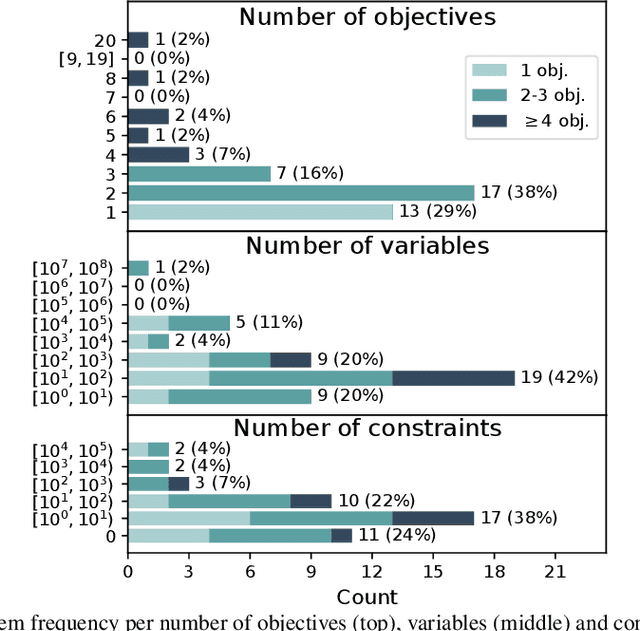
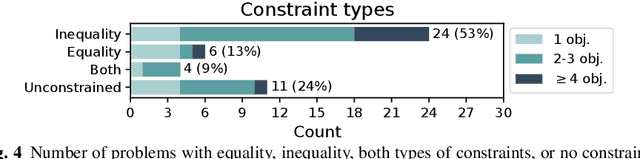
Abstract:Optimisation algorithms are commonly compared on benchmarks to get insight into performance differences. However, it is not clear how closely benchmarks match the properties of real-world problems because these properties are largely unknown. This work investigates the properties of real-world problems through a questionnaire to enable the design of future benchmark problems that more closely resemble those found in the real world. The results, while not representative, show that many problems possess at least one of the following properties: they are constrained, deterministic, have only continuous variables, require substantial computation times for both the objectives and the constraints, or allow a limited number of evaluations. Properties like known optimal solutions and analytical gradients are rarely available, limiting the options in guiding the optimisation process. These are all important aspects to consider when designing realistic benchmark problems. At the same time, objective functions are often reported to be black-box and since many problem properties are unknown the design of realistic benchmarks is difficult. To further improve the understanding of real-world problems, readers working on a real-world optimisation problem are encouraged to fill out the questionnaire: https://tinyurl.com/opt-survey
Towards Realistic Optimization Benchmarks: A Questionnaire on the Properties of Real-World Problems
Apr 14, 2020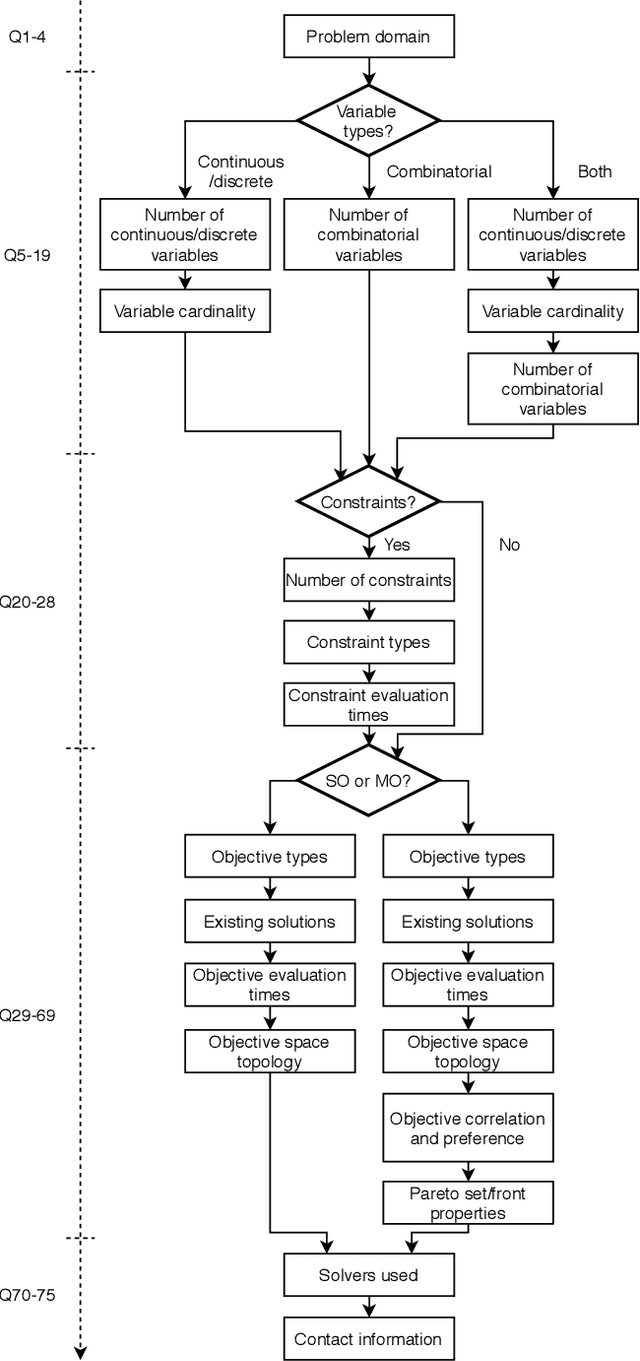
Abstract:Benchmarks are a useful tool for empirical performance comparisons. However, one of the main shortcomings of existing benchmarks is that it remains largely unclear how they relate to real-world problems. What does an algorithm's performance on a benchmark say about its potential on a specific real-world problem? This work aims to identify properties of real-world problems through a questionnaire on real-world single-, multi-, and many-objective optimization problems. Based on initial responses, a few challenges that have to be considered in the design of realistic benchmarks can already be identified. A key point for future work is to gather more responses to the questionnaire to allow an analysis of common combinations of properties. In turn, such common combinations can then be included in improved benchmark suites. To gather more data, the reader is invited to participate in the questionnaire at: https://tinyurl.com/opt-survey
An End-to-end Deep Learning Approach for Landmark Detection and Matching in Medical Images
Jan 21, 2020



Abstract:Anatomical landmark correspondences in medical images can provide additional guidance information for the alignment of two images, which, in turn, is crucial for many medical applications. However, manual landmark annotation is labor-intensive. Therefore, we propose an end-to-end deep learning approach to automatically detect landmark correspondences in pairs of two-dimensional (2D) images. Our approach consists of a Siamese neural network, which is trained to identify salient locations in images as landmarks and predict matching probabilities for landmark pairs from two different images. We trained our approach on 2D transverse slices from 168 lower abdominal Computed Tomography (CT) scans. We tested the approach on 22,206 pairs of 2D slices with varying levels of intensity, affine, and elastic transformations. The proposed approach finds an average of 639, 466, and 370 landmark matches per image pair for intensity, affine, and elastic transformations, respectively, with spatial matching errors of at most 1 mm. Further, more than 99% of the landmark pairs are within a spatial matching error of 2 mm, 4 mm, and 8 mm for image pairs with intensity, affine, and elastic transformations, respectively. To investigate the utility of our developed approach in a clinical setting, we also tested our approach on pairs of transverse slices selected from follow-up CT scans of three patients. Visual inspection of the results revealed landmark matches in both bony anatomical regions as well as in soft tissues lacking prominent intensity gradients.
A Drug Recommendation System for cancer cell lines
Dec 24, 2019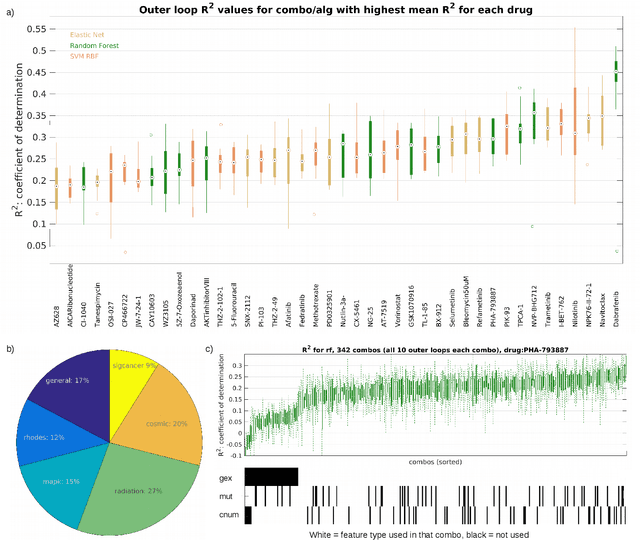
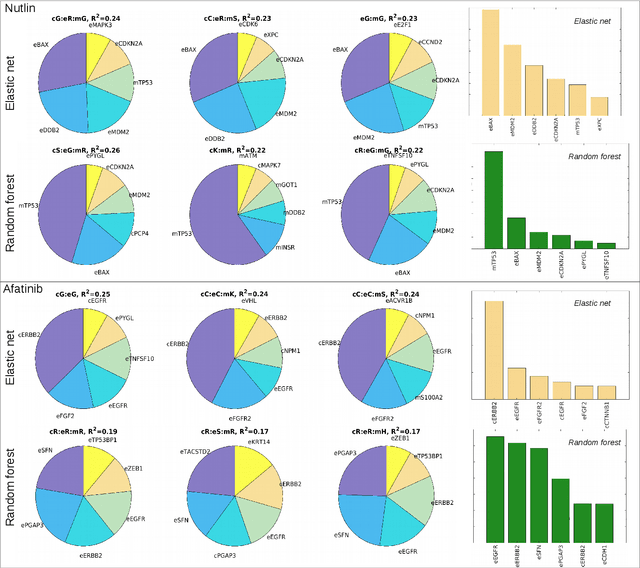
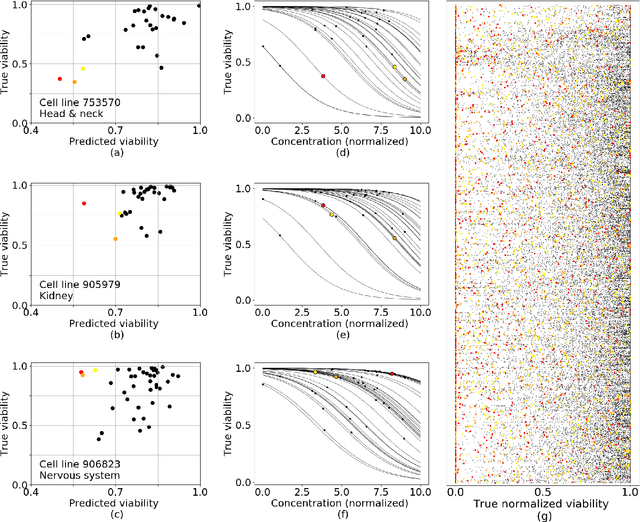
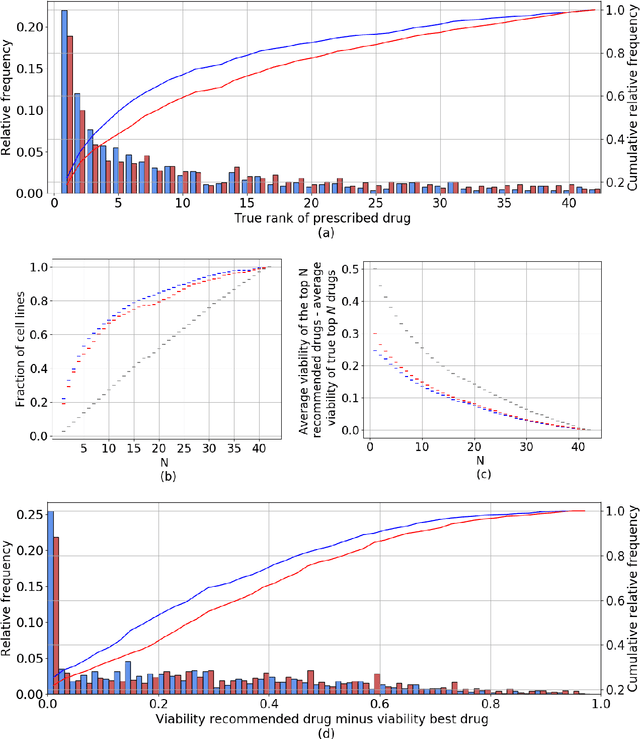
Abstract:Personalizing drug prescriptions in cancer care based on genomic information requires associating genomic markers with treatment effects. This is an unsolved challenge requiring genomic patient data in yet unavailable volumes as well as appropriate quantitative methods. We attempt to solve this challenge for an experimental proxy for which sufficient data is available: 42 drugs tested on 1018 cancer cell lines. Our goal is to develop a method to identify the drug that is most promising based on a cell line's genomic information. For this, we need to identify for each drug the machine learning method, choice of hyperparameters and genomic features for optimal predictive performance. We extensively compare combinations of gene sets (both curated and random), genetic features, and machine learning algorithms for all 42 drugs. For each drug, the best performing combination (considering only the curated gene sets) is selected. We use these top model parameters for each drug to build and demonstrate a Drug Recommendation System (Dr.S). Insights resulting from this analysis are formulated as best practices for developing drug recommendation systems. The complete software system, called the Cell Line Analyzer, is written in Python and available on github.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge