Jan Wiersma
Sharing Knowledge without Sharing Data: Stitches can improve ensembles of disjointly trained models
Dec 19, 2025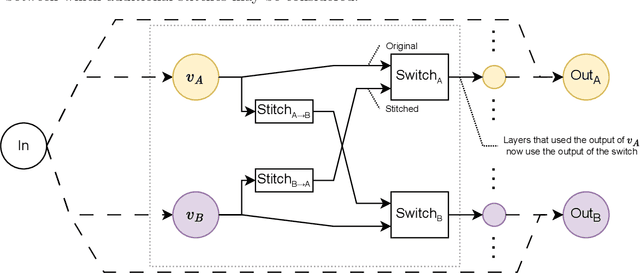
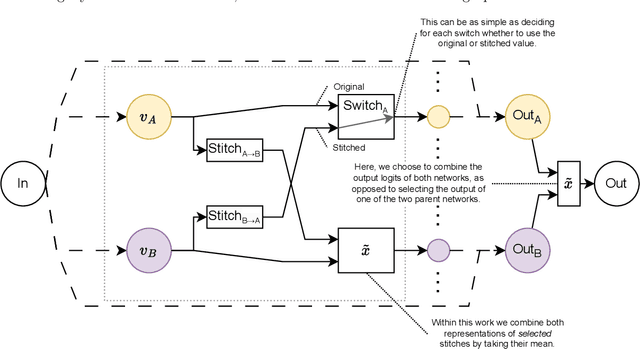
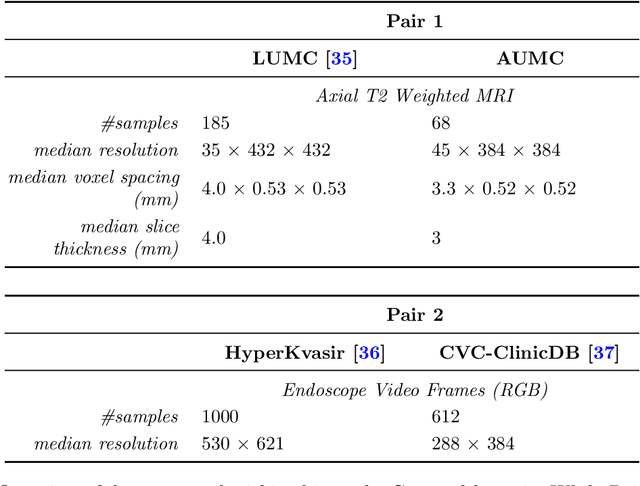
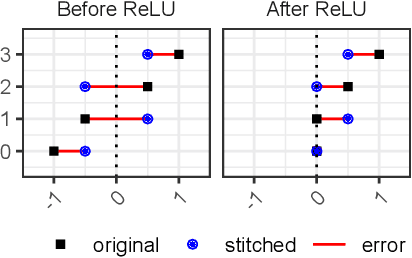
Abstract:Deep learning has been shown to be very capable at performing many real-world tasks. However, this performance is often dependent on the presence of large and varied datasets. In some settings, like in the medical domain, data is often fragmented across parties, and cannot be readily shared. While federated learning addresses this situation, it is a solution that requires synchronicity of parties training a single model together, exchanging information about model weights. We investigate how asynchronous collaboration, where only already trained models are shared (e.g. as part of a publication), affects performance, and propose to use stitching as a method for combining models. Through taking a multi-objective perspective, where performance on each parties' data is viewed independently, we find that training solely on a single parties' data results in similar performance when merging with another parties' data, when considering performance on that single parties' data, while performance on other parties' data is notably worse. Moreover, while an ensemble of such individually trained networks generalizes better, performance on each parties' own dataset suffers. We find that combining intermediate representations in individually trained models with a well placed pair of stitching layers allows this performance to recover to a competitive degree while maintaining improved generalization, showing that asynchronous collaboration can yield competitive results.
Automatic Landmarks Correspondence Detection in Medical Images with an Application to Deformable Image Registration
Sep 06, 2021



Abstract:Deformable Image Registration (DIR) can benefit from additional guidance using corresponding landmarks in the images. However, the benefits thereof are largely understudied, especially due to the lack of automatic detection methods for corresponding landmarks in three-dimensional (3D) medical images. In this work, we present a Deep Convolutional Neural Network (DCNN), called DCNN-Match, that learns to predict landmark correspondences in 3D images in a self-supervised manner. We explored five variants of DCNN-Match that use different loss functions and tested DCNN-Match separately as well as in combination with the open-source registration software Elastix to assess its impact on a common DIR approach. We employed lower-abdominal Computed Tomography (CT) scans from cervical cancer patients: 121 pelvic CT scan pairs containing simulated elastic transformations and 11 pairs demonstrating clinical deformations. Our results show significant improvement in DIR performance when landmark correspondences predicted by DCNN-Match were used in case of simulated as well as clinical deformations. We also observed that the spatial distribution of the automatically identified landmarks and the associated matching errors affect the extent of improvement in DIR. Finally, DCNN-Match was found to generalize well to Magnetic Resonance Imaging (MRI) scans without requiring retraining, indicating easy applicability to other datasets.
An End-to-end Deep Learning Approach for Landmark Detection and Matching in Medical Images
Jan 21, 2020



Abstract:Anatomical landmark correspondences in medical images can provide additional guidance information for the alignment of two images, which, in turn, is crucial for many medical applications. However, manual landmark annotation is labor-intensive. Therefore, we propose an end-to-end deep learning approach to automatically detect landmark correspondences in pairs of two-dimensional (2D) images. Our approach consists of a Siamese neural network, which is trained to identify salient locations in images as landmarks and predict matching probabilities for landmark pairs from two different images. We trained our approach on 2D transverse slices from 168 lower abdominal Computed Tomography (CT) scans. We tested the approach on 22,206 pairs of 2D slices with varying levels of intensity, affine, and elastic transformations. The proposed approach finds an average of 639, 466, and 370 landmark matches per image pair for intensity, affine, and elastic transformations, respectively, with spatial matching errors of at most 1 mm. Further, more than 99% of the landmark pairs are within a spatial matching error of 2 mm, 4 mm, and 8 mm for image pairs with intensity, affine, and elastic transformations, respectively. To investigate the utility of our developed approach in a clinical setting, we also tested our approach on pairs of transverse slices selected from follow-up CT scans of three patients. Visual inspection of the results revealed landmark matches in both bony anatomical regions as well as in soft tissues lacking prominent intensity gradients.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge