Alexander Jaus
Semantic Segmentation for Preoperative Planning in Transcatheter Aortic Valve Replacement
Jul 22, 2025Abstract:When preoperative planning for surgeries is conducted on the basis of medical images, artificial intelligence methods can support medical doctors during assessment. In this work, we consider medical guidelines for preoperative planning of the transcatheter aortic valve replacement (TAVR) and identify tasks, that may be supported via semantic segmentation models by making relevant anatomical structures measurable in computed tomography scans. We first derive fine-grained TAVR-relevant pseudo-labels from coarse-grained anatomical information, in order to train segmentation models and quantify how well they are able to find these structures in the scans. Furthermore, we propose an adaptation to the loss function in training these segmentation models and through this achieve a +1.27% Dice increase in performance. Our fine-grained TAVR-relevant pseudo-labels and the computed tomography scans we build upon are available at https://doi.org/10.5281/zenodo.16274176.
Is Visual in-Context Learning for Compositional Medical Tasks within Reach?
Jul 02, 2025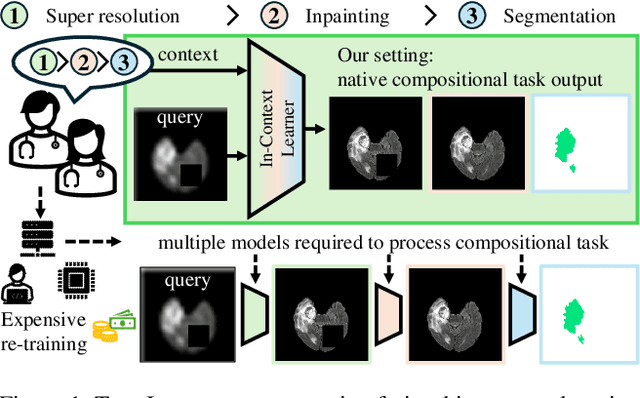
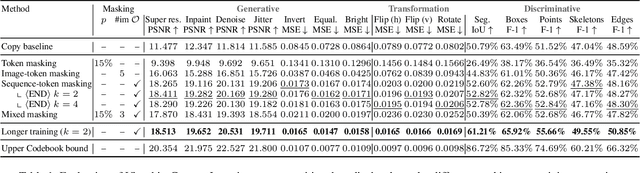
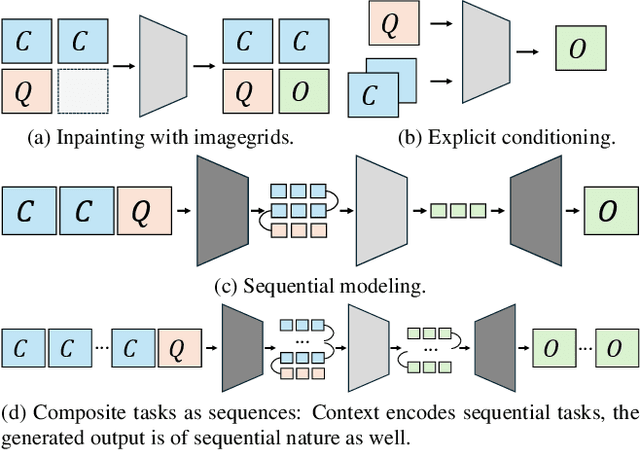
Abstract:In this paper, we explore the potential of visual in-context learning to enable a single model to handle multiple tasks and adapt to new tasks during test time without re-training. Unlike previous approaches, our focus is on training in-context learners to adapt to sequences of tasks, rather than individual tasks. Our goal is to solve complex tasks that involve multiple intermediate steps using a single model, allowing users to define entire vision pipelines flexibly at test time. To achieve this, we first examine the properties and limitations of visual in-context learning architectures, with a particular focus on the role of codebooks. We then introduce a novel method for training in-context learners using a synthetic compositional task generation engine. This engine bootstraps task sequences from arbitrary segmentation datasets, enabling the training of visual in-context learners for compositional tasks. Additionally, we investigate different masking-based training objectives to gather insights into how to train models better for solving complex, compositional tasks. Our exploration not only provides important insights especially for multi-modal medical task sequences but also highlights challenges that need to be addressed.
Good Enough: Is it Worth Improving your Label Quality?
May 27, 2025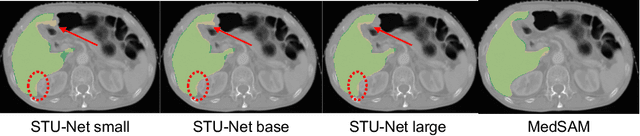
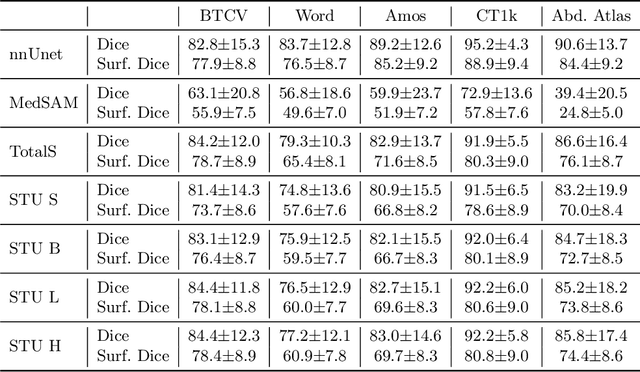
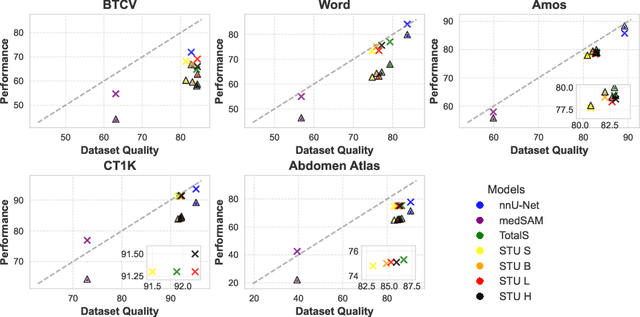
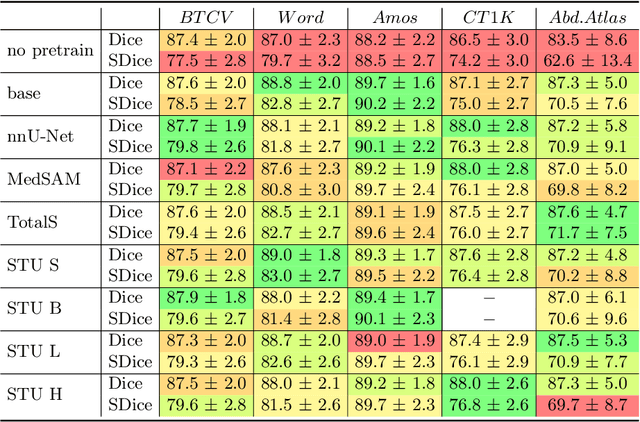
Abstract:Improving label quality in medical image segmentation is costly, but its benefits remain unclear. We systematically evaluate its impact using multiple pseudo-labeled versions of CT datasets, generated by models like nnU-Net, TotalSegmentator, and MedSAM. Our results show that while higher-quality labels improve in-domain performance, gains remain unclear if below a small threshold. For pre-training, label quality has minimal impact, suggesting that models rather transfer general concepts than detailed annotations. These findings provide guidance on when improving label quality is worth the effort.
Muscles in Time: Learning to Understand Human Motion by Simulating Muscle Activations
Oct 31, 2024



Abstract:Exploring the intricate dynamics between muscular and skeletal structures is pivotal for understanding human motion. This domain presents substantial challenges, primarily attributed to the intensive resources required for acquiring ground truth muscle activation data, resulting in a scarcity of datasets. In this work, we address this issue by establishing Muscles in Time (MinT), a large-scale synthetic muscle activation dataset. For the creation of MinT, we enriched existing motion capture datasets by incorporating muscle activation simulations derived from biomechanical human body models using the OpenSim platform, a common approach in biomechanics and human motion research. Starting from simple pose sequences, our pipeline enables us to extract detailed information about the timing of muscle activations within the human musculoskeletal system. Muscles in Time contains over nine hours of simulation data covering 227 subjects and 402 simulated muscle strands. We demonstrate the utility of this dataset by presenting results on neural network-based muscle activation estimation from human pose sequences with two different sequence-to-sequence architectures. Data and code are provided under https://simplexsigil.github.io/mint.
Every Component Counts: Rethinking the Measure of Success for Medical Semantic Segmentation in Multi-Instance Segmentation Tasks
Oct 24, 2024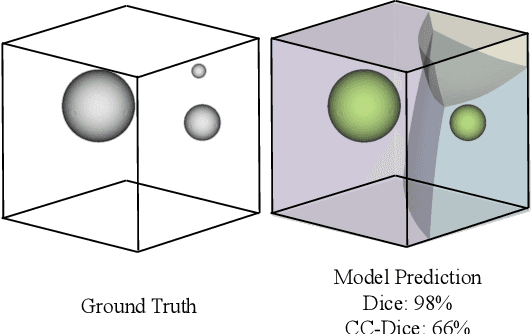
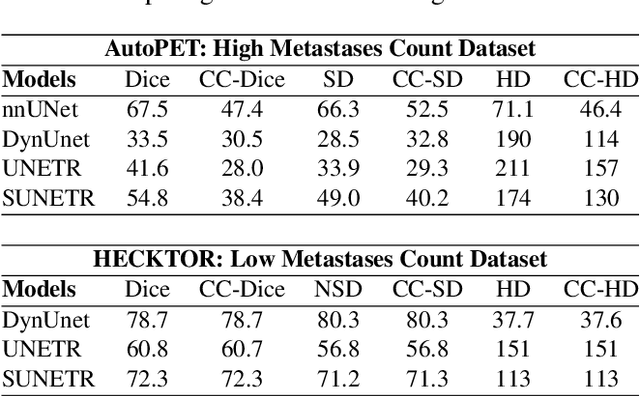
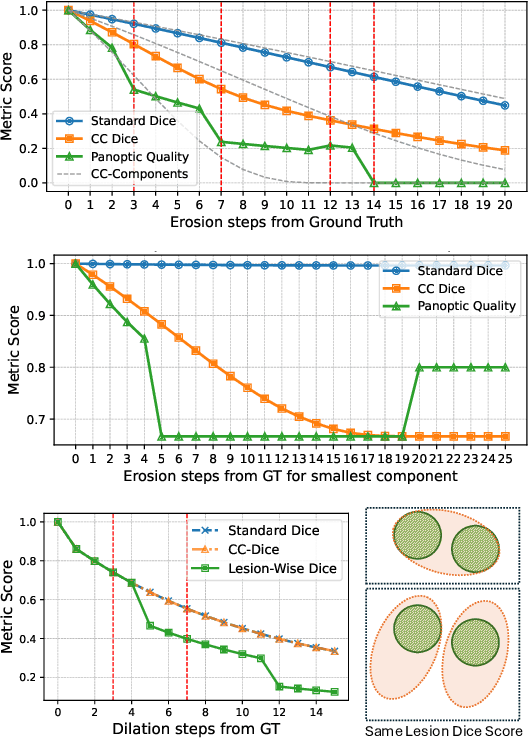
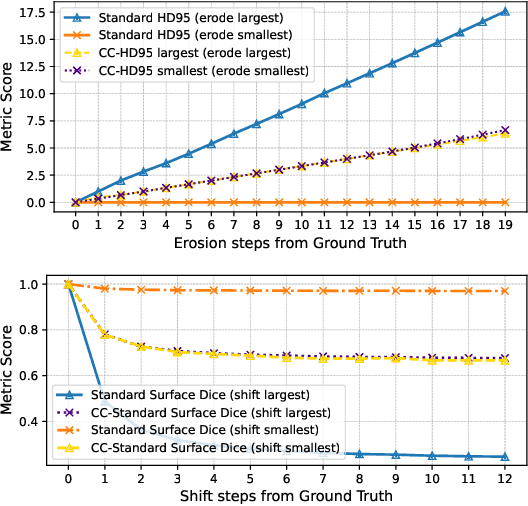
Abstract:We present Connected-Component~(CC)-Metrics, a novel semantic segmentation evaluation protocol, targeted to align existing semantic segmentation metrics to a multi-instance detection scenario in which each connected component matters. We motivate this setup in the common medical scenario of semantic metastases segmentation in a full-body PET/CT. We show how existing semantic segmentation metrics suffer from a bias towards larger connected components contradicting the clinical assessment of scans in which tumor size and clinical relevance are uncorrelated. To rebalance existing segmentation metrics, we propose to evaluate them on a per-component basis thus giving each tumor the same weight irrespective of its size. To match predictions to ground-truth segments, we employ a proximity-based matching criterion, evaluating common metrics locally at the component of interest. Using this approach, we break free of biases introduced by large metastasis for overlap-based metrics such as Dice or Surface Dice. CC-Metrics also improves distance-based metrics such as Hausdorff Distances which are uninformative for small changes that do not influence the maximum or 95th percentile, and avoids pitfalls introduced by directly combining counting-based metrics with overlap-based metrics as it is done in Panoptic Quality.
LIMIS: Towards Language-based Interactive Medical Image Segmentation
Oct 22, 2024Abstract:Within this work, we introduce LIMIS: The first purely language-based interactive medical image segmentation model. We achieve this by adapting Grounded SAM to the medical domain and designing a language-based model interaction strategy that allows radiologists to incorporate their knowledge into the segmentation process. LIMIS produces high-quality initial segmentation masks by leveraging medical foundation models and allows users to adapt segmentation masks using only language, opening up interactive segmentation to scenarios where physicians require using their hands for other tasks. We evaluate LIMIS on three publicly available medical datasets in terms of performance and usability with experts from the medical domain confirming its high-quality segmentation masks and its interactive usability.
Anatomy-guided Pathology Segmentation
Jul 08, 2024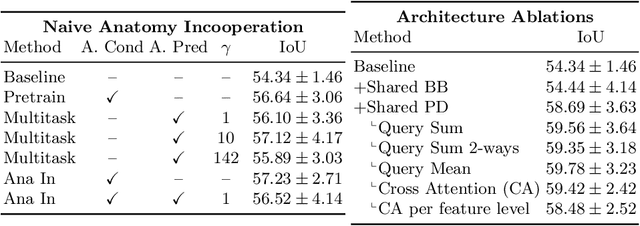
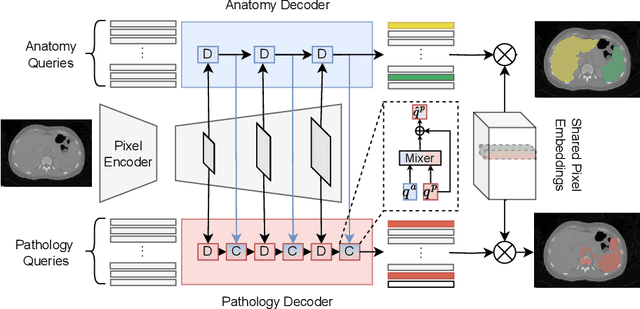
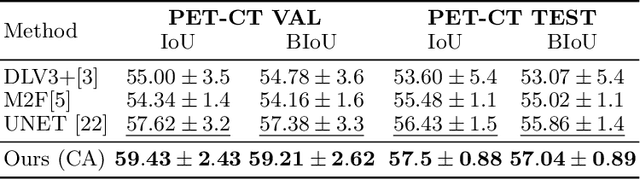
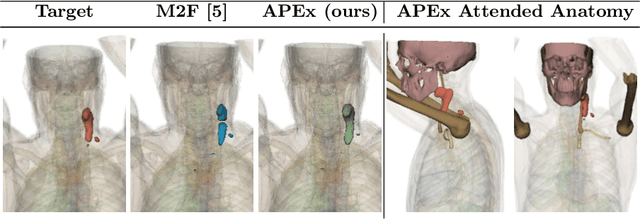
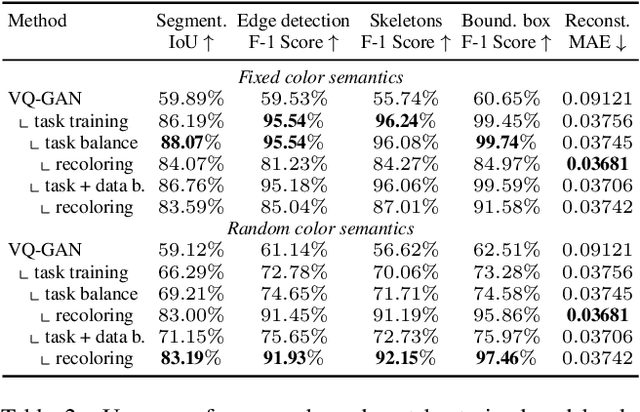
Abstract:Pathological structures in medical images are typically deviations from the expected anatomy of a patient. While clinicians consider this interplay between anatomy and pathology, recent deep learning algorithms specialize in recognizing either one of the two, rarely considering the patient's body from such a joint perspective. In this paper, we develop a generalist segmentation model that combines anatomical and pathological information, aiming to enhance the segmentation accuracy of pathological features. Our Anatomy-Pathology Exchange (APEx) training utilizes a query-based segmentation transformer which decodes a joint feature space into query-representations for human anatomy and interleaves them via a mixing strategy into the pathology-decoder for anatomy-informed pathology predictions. In doing so, we are able to report the best results across the board on FDG-PET-CT and Chest X-Ray pathology segmentation tasks with a margin of up to 3.3% as compared to strong baseline methods. Code and models will be publicly available at github.com/alexanderjaus/APEx.
MedShapeNet -- A Large-Scale Dataset of 3D Medical Shapes for Computer Vision
Sep 12, 2023



Abstract:We present MedShapeNet, a large collection of anatomical shapes (e.g., bones, organs, vessels) and 3D surgical instrument models. Prior to the deep learning era, the broad application of statistical shape models (SSMs) in medical image analysis is evidence that shapes have been commonly used to describe medical data. Nowadays, however, state-of-the-art (SOTA) deep learning algorithms in medical imaging are predominantly voxel-based. In computer vision, on the contrary, shapes (including, voxel occupancy grids, meshes, point clouds and implicit surface models) are preferred data representations in 3D, as seen from the numerous shape-related publications in premier vision conferences, such as the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), as well as the increasing popularity of ShapeNet (about 51,300 models) and Princeton ModelNet (127,915 models) in computer vision research. MedShapeNet is created as an alternative to these commonly used shape benchmarks to facilitate the translation of data-driven vision algorithms to medical applications, and it extends the opportunities to adapt SOTA vision algorithms to solve critical medical problems. Besides, the majority of the medical shapes in MedShapeNet are modeled directly on the imaging data of real patients, and therefore it complements well existing shape benchmarks comprising of computer-aided design (CAD) models. MedShapeNet currently includes more than 100,000 medical shapes, and provides annotations in the form of paired data. It is therefore also a freely available repository of 3D models for extended reality (virtual reality - VR, augmented reality - AR, mixed reality - MR) and medical 3D printing. This white paper describes in detail the motivations behind MedShapeNet, the shape acquisition procedures, the use cases, as well as the usage of the online shape search portal: https://medshapenet.ikim.nrw/
Towards Unifying Anatomy Segmentation: Automated Generation of a Full-body CT Dataset via Knowledge Aggregation and Anatomical Guidelines
Jul 25, 2023Abstract:In this study, we present a method for generating automated anatomy segmentation datasets using a sequential process that involves nnU-Net-based pseudo-labeling and anatomy-guided pseudo-label refinement. By combining various fragmented knowledge bases, we generate a dataset of whole-body CT scans with $142$ voxel-level labels for 533 volumes providing comprehensive anatomical coverage which experts have approved. Our proposed procedure does not rely on manual annotation during the label aggregation stage. We examine its plausibility and usefulness using three complementary checks: Human expert evaluation which approved the dataset, a Deep Learning usefulness benchmark on the BTCV dataset in which we achieve 85% dice score without using its training dataset, and medical validity checks. This evaluation procedure combines scalable automated checks with labor-intensive high-quality expert checks. Besides the dataset, we release our trained unified anatomical segmentation model capable of predicting $142$ anatomical structures on CT data.
Accurate Fine-Grained Segmentation of Human Anatomy in Radiographs via Volumetric Pseudo-Labeling
Jun 06, 2023Abstract:Purpose: Interpreting chest radiographs (CXR) remains challenging due to the ambiguity of overlapping structures such as the lungs, heart, and bones. To address this issue, we propose a novel method for extracting fine-grained anatomical structures in CXR using pseudo-labeling of three-dimensional computed tomography (CT) scans. Methods: We created a large-scale dataset of 10,021 thoracic CTs with 157 labels and applied an ensemble of 3D anatomy segmentation models to extract anatomical pseudo-labels. These labels were projected onto a two-dimensional plane, similar to the CXR, allowing the training of detailed semantic segmentation models for CXR without any manual annotation effort. Results: Our resulting segmentation models demonstrated remarkable performance on CXR, with a high average model-annotator agreement between two radiologists with mIoU scores of 0.93 and 0.85 for frontal and lateral anatomy, while inter-annotator agreement remained at 0.95 and 0.83 mIoU. Our anatomical segmentations allowed for the accurate extraction of relevant explainable medical features such as the cardio-thoracic-ratio. Conclusion: Our method of volumetric pseudo-labeling paired with CT projection offers a promising approach for detailed anatomical segmentation of CXR with a high agreement with human annotators. This technique may have important clinical implications, particularly in the analysis of various thoracic pathologies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge