Michael Kamp
CISPA Helmholtz Center for Information Security
When Flatness Does (Not) Guarantee Adversarial Robustness
Oct 16, 2025Abstract:Despite their empirical success, neural networks remain vulnerable to small, adversarial perturbations. A longstanding hypothesis suggests that flat minima, regions of low curvature in the loss landscape, offer increased robustness. While intuitive, this connection has remained largely informal and incomplete. By rigorously formalizing the relationship, we show this intuition is only partially correct: flatness implies local but not global adversarial robustness. To arrive at this result, we first derive a closed-form expression for relative flatness in the penultimate layer, and then show we can use this to constrain the variation of the loss in input space. This allows us to formally analyze the adversarial robustness of the entire network. We then show that to maintain robustness beyond a local neighborhood, the loss needs to curve sharply away from the data manifold. We validate our theoretical predictions empirically across architectures and datasets, uncovering the geometric structure that governs adversarial vulnerability, and linking flatness to model confidence: adversarial examples often lie in large, flat regions where the model is confidently wrong. Our results challenge simplified views of flatness and provide a nuanced understanding of its role in robustness.
Federated Binary Matrix Factorization using Proximal Optimization
Jul 01, 2024Abstract:Identifying informative components in binary data is an essential task in many research areas, including life sciences, social sciences, and recommendation systems. Boolean matrix factorization (BMF) is a family of methods that performs this task by efficiently factorizing the data. In real-world settings, the data is often distributed across stakeholders and required to stay private, prohibiting the straightforward application of BMF. To adapt BMF to this context, we approach the problem from a federated-learning perspective, while building on a state-of-the-art continuous binary matrix factorization relaxation to BMF that enables efficient gradient-based optimization. We propose to only share the relaxed component matrices, which are aggregated centrally using a proximal operator that regularizes for binary outcomes. We show the convergence of our federated proximal gradient descent algorithm and provide differential privacy guarantees. Our extensive empirical evaluation demonstrates that our algorithm outperforms, in terms of quality and efficacy, federation schemes of state-of-the-art BMF methods on a diverse set of real-world and synthetic data.
Landscaping Linear Mode Connectivity
Jun 24, 2024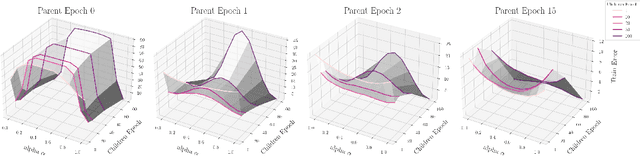
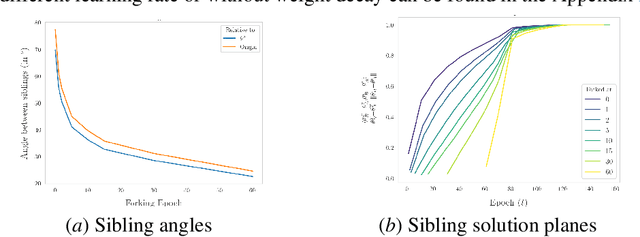
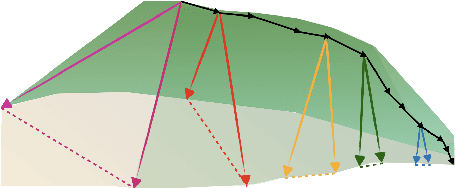
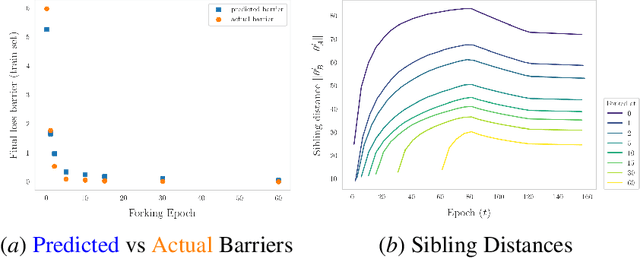
Abstract:The presence of linear paths in parameter space between two different network solutions in certain cases, i.e., linear mode connectivity (LMC), has garnered interest from both theoretical and practical fronts. There has been significant research that either practically designs algorithms catered for connecting networks by adjusting for the permutation symmetries as well as some others that more theoretically construct paths through which networks can be connected. Yet, the core reasons for the occurrence of LMC, when in fact it does occur, in the highly non-convex loss landscapes of neural networks are far from clear. In this work, we take a step towards understanding it by providing a model of how the loss landscape needs to behave topographically for LMC (or the lack thereof) to manifest. Concretely, we present a `mountainside and ridge' perspective that helps to neatly tie together different geometric features that can be spotted in the loss landscape along the training runs. We also complement this perspective by providing a theoretical analysis of the barrier height, for which we provide empirical support, and which additionally extends as a faithful predictor of layer-wise LMC. We close with a toy example that provides further intuition on how barriers arise in the first place, all in all, showcasing the larger aim of the work -- to provide a working model of the landscape and its topography for the occurrence of LMC.
The Uncanny Valley: Exploring Adversarial Robustness from a Flatness Perspective
May 27, 2024



Abstract:Flatness of the loss surface not only correlates positively with generalization but is also related to adversarial robustness, since perturbations of inputs relate non-linearly to perturbations of weights. In this paper, we empirically analyze the relation between adversarial examples and relative flatness with respect to the parameters of one layer. We observe a peculiar property of adversarial examples: during an iterative first-order white-box attack, the flatness of the loss surface measured around the adversarial example first becomes sharper until the label is flipped, but if we keep the attack running it runs into a flat uncanny valley where the label remains flipped. We find this phenomenon across various model architectures and datasets. Our results also extend to large language models (LLMs), but due to the discrete nature of the input space and comparatively weak attacks, the adversarial examples rarely reach a truly flat region. Most importantly, this phenomenon shows that flatness alone cannot explain adversarial robustness unless we can also guarantee the behavior of the function around the examples. We theoretically connect relative flatness to adversarial robustness by bounding the third derivative of the loss surface, underlining the need for flatness in combination with a low global Lipschitz constant for a robust model.
Orthogonal Gradient Boosting for Simpler Additive Rule Ensembles
Feb 24, 2024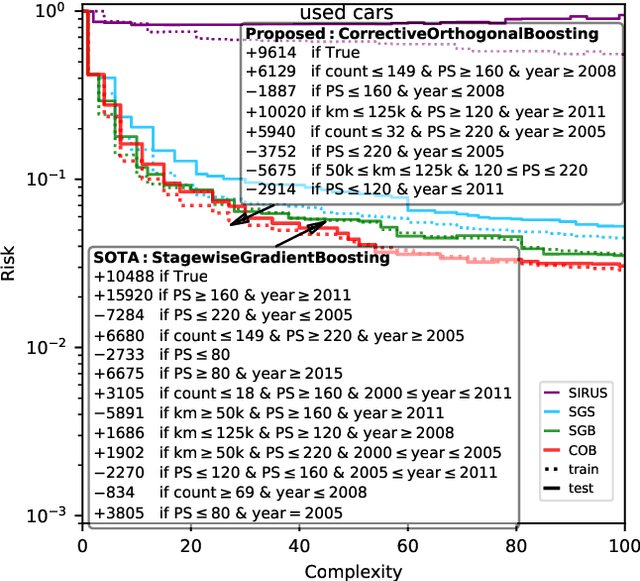
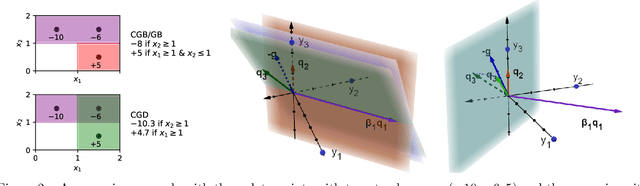
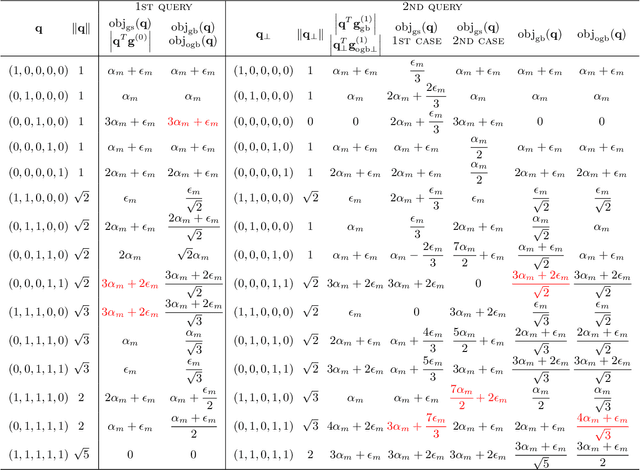
Abstract:Gradient boosting of prediction rules is an efficient approach to learn potentially interpretable yet accurate probabilistic models. However, actual interpretability requires to limit the number and size of the generated rules, and existing boosting variants are not designed for this purpose. Though corrective boosting refits all rule weights in each iteration to minimise prediction risk, the included rule conditions tend to be sub-optimal, because commonly used objective functions fail to anticipate this refitting. Here, we address this issue by a new objective function that measures the angle between the risk gradient vector and the projection of the condition output vector onto the orthogonal complement of the already selected conditions. This approach correctly approximate the ideal update of adding the risk gradient itself to the model and favours the inclusion of more general and thus shorter rules. As we demonstrate using a wide range of prediction tasks, this significantly improves the comprehensibility/accuracy trade-off of the fitted ensemble. Additionally, we show how objective values for related rule conditions can be computed incrementally to avoid any substantial computational overhead of the new method.
Protecting Sensitive Data through Federated Co-Training
Oct 09, 2023



Abstract:In many critical applications, sensitive data is inherently distributed. Federated learning trains a model collaboratively by aggregating the parameters of locally trained models. This avoids exposing sensitive local data. It is possible, though, to infer upon the sensitive data from the shared model parameters. At the same time, many types of machine learning models do not lend themselves to parameter aggregation, such as decision trees, or rule ensembles. It has been observed that in many applications, in particular healthcare, large unlabeled datasets are publicly available. They can be used to exchange information between clients by distributed distillation, i.e., co-regularizing local training via the discrepancy between the soft predictions of each local client on the unlabeled dataset. This, however, still discloses private information and restricts the types of models to those trainable via gradient-based methods. We propose to go one step further and use a form of federated co-training, where local hard labels on the public unlabeled datasets are shared and aggregated into a consensus label. This consensus label can be used for local training by any supervised machine learning model. We show that this federated co-training approach achieves a model quality comparable to both federated learning and distributed distillation on a set of benchmark datasets and real-world medical datasets. It improves privacy over both approaches, protecting against common membership inference attacks to the highest degree. Furthermore, we show that federated co-training can collaboratively train interpretable models, such as decision trees and rule ensembles, achieving a model quality comparable to centralized training.
MedShapeNet -- A Large-Scale Dataset of 3D Medical Shapes for Computer Vision
Sep 12, 2023



Abstract:We present MedShapeNet, a large collection of anatomical shapes (e.g., bones, organs, vessels) and 3D surgical instrument models. Prior to the deep learning era, the broad application of statistical shape models (SSMs) in medical image analysis is evidence that shapes have been commonly used to describe medical data. Nowadays, however, state-of-the-art (SOTA) deep learning algorithms in medical imaging are predominantly voxel-based. In computer vision, on the contrary, shapes (including, voxel occupancy grids, meshes, point clouds and implicit surface models) are preferred data representations in 3D, as seen from the numerous shape-related publications in premier vision conferences, such as the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), as well as the increasing popularity of ShapeNet (about 51,300 models) and Princeton ModelNet (127,915 models) in computer vision research. MedShapeNet is created as an alternative to these commonly used shape benchmarks to facilitate the translation of data-driven vision algorithms to medical applications, and it extends the opportunities to adapt SOTA vision algorithms to solve critical medical problems. Besides, the majority of the medical shapes in MedShapeNet are modeled directly on the imaging data of real patients, and therefore it complements well existing shape benchmarks comprising of computer-aided design (CAD) models. MedShapeNet currently includes more than 100,000 medical shapes, and provides annotations in the form of paired data. It is therefore also a freely available repository of 3D models for extended reality (virtual reality - VR, augmented reality - AR, mixed reality - MR) and medical 3D printing. This white paper describes in detail the motivations behind MedShapeNet, the shape acquisition procedures, the use cases, as well as the usage of the online shape search portal: https://medshapenet.ikim.nrw/
FAM: Relative Flatness Aware Minimization
Jul 05, 2023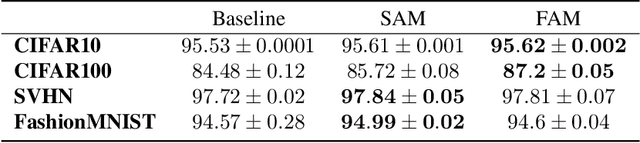
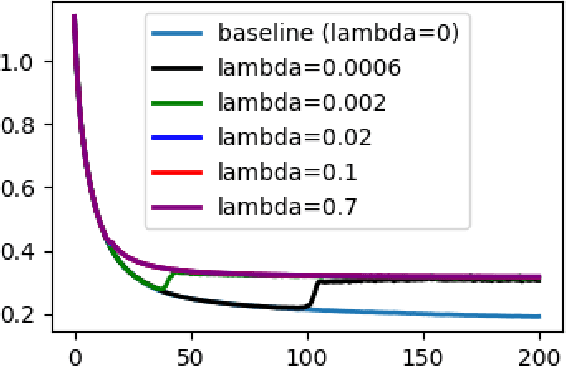
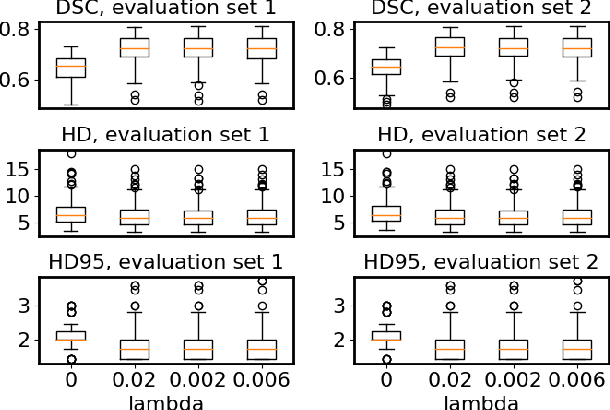
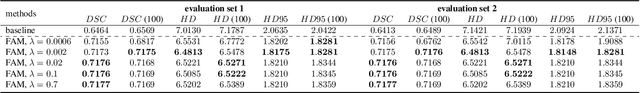
Abstract:Flatness of the loss curve around a model at hand has been shown to empirically correlate with its generalization ability. Optimizing for flatness has been proposed as early as 1994 by Hochreiter and Schmidthuber, and was followed by more recent successful sharpness-aware optimization techniques. Their widespread adoption in practice, though, is dubious because of the lack of theoretically grounded connection between flatness and generalization, in particular in light of the reparameterization curse - certain reparameterizations of a neural network change most flatness measures but do not change generalization. Recent theoretical work suggests that a particular relative flatness measure can be connected to generalization and solves the reparameterization curse. In this paper, we derive a regularizer based on this relative flatness that is easy to compute, fast, efficient, and works with arbitrary loss functions. It requires computing the Hessian only of a single layer of the network, which makes it applicable to large neural networks, and with it avoids an expensive mapping of the loss surface in the vicinity of the model. In an extensive empirical evaluation we show that this relative flatness aware minimization (FAM) improves generalization in a multitude of applications and models, both in finetuning and standard training. We make the code available at github.
Why does my medical AI look at pictures of birds? Exploring the efficacy of transfer learning across domain boundaries
Jun 30, 2023Abstract:It is an open secret that ImageNet is treated as the panacea of pretraining. Particularly in medical machine learning, models not trained from scratch are often finetuned based on ImageNet-pretrained models. We posit that pretraining on data from the domain of the downstream task should almost always be preferred instead. We leverage RadNet-12M, a dataset containing more than 12 million computed tomography (CT) image slices, to explore the efficacy of self-supervised pretraining on medical and natural images. Our experiments cover intra- and cross-domain transfer scenarios, varying data scales, finetuning vs. linear evaluation, and feature space analysis. We observe that intra-domain transfer compares favorably to cross-domain transfer, achieving comparable or improved performance (0.44% - 2.07% performance increase using RadNet pretraining, depending on the experiment) and demonstrate the existence of a domain boundary-related generalization gap and domain-specific learned features.
Open-Source Skull Reconstruction with MONAI
Nov 25, 2022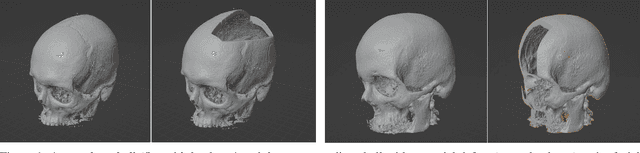
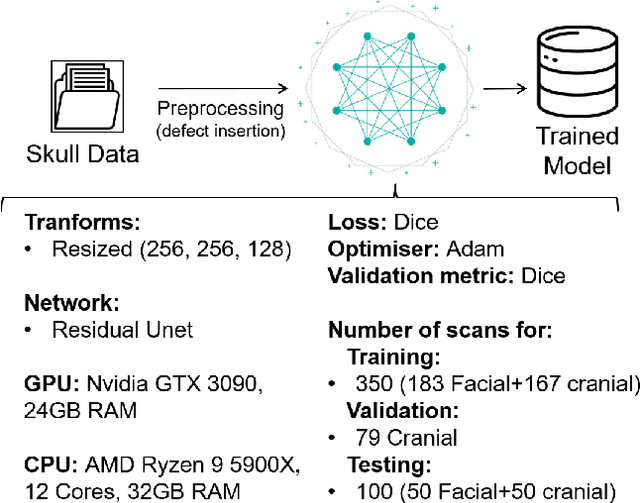
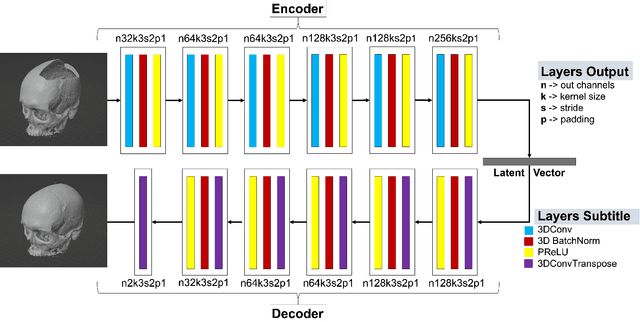
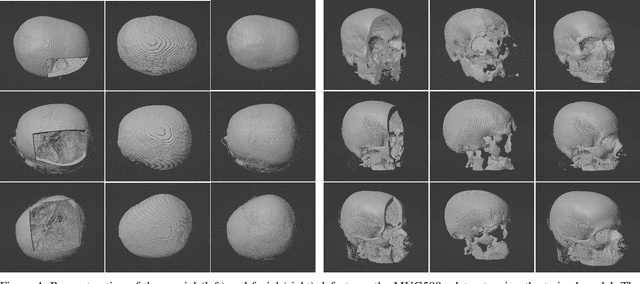
Abstract:We present a deep learning-based approach for skull reconstruction for MONAI, which has been pre-trained on the MUG500+ skull dataset. The implementation follows the MONAI contribution guidelines, hence, it can be easily tried out and used, and extended by MONAI users. The primary goal of this paper lies in the investigation of open-sourcing codes and pre-trained deep learning models under the MONAI framework. Nowadays, open-sourcing software, especially (pre-trained) deep learning models, has become increasingly important. Over the years, medical image analysis experienced a tremendous transformation. Over a decade ago, algorithms had to be implemented and optimized with low-level programming languages, like C or C++, to run in a reasonable time on a desktop PC, which was not as powerful as today's computers. Nowadays, users have high-level scripting languages like Python, and frameworks like PyTorch and TensorFlow, along with a sea of public code repositories at hand. As a result, implementations that had thousands of lines of C or C++ code in the past, can now be scripted with a few lines and in addition executed in a fraction of the time. To put this even on a higher level, the Medical Open Network for Artificial Intelligence (MONAI) framework tailors medical imaging research to an even more convenient process, which can boost and push the whole field. The MONAI framework is a freely available, community-supported, open-source and PyTorch-based framework, that also enables to provide research contributions with pre-trained models to others. Codes and pre-trained weights for skull reconstruction are publicly available at: https://github.com/Project-MONAI/research-contributions/tree/master/SkullRec
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge