Moon-Sung Kim
MedShapeNet -- A Large-Scale Dataset of 3D Medical Shapes for Computer Vision
Sep 12, 2023



Abstract:We present MedShapeNet, a large collection of anatomical shapes (e.g., bones, organs, vessels) and 3D surgical instrument models. Prior to the deep learning era, the broad application of statistical shape models (SSMs) in medical image analysis is evidence that shapes have been commonly used to describe medical data. Nowadays, however, state-of-the-art (SOTA) deep learning algorithms in medical imaging are predominantly voxel-based. In computer vision, on the contrary, shapes (including, voxel occupancy grids, meshes, point clouds and implicit surface models) are preferred data representations in 3D, as seen from the numerous shape-related publications in premier vision conferences, such as the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), as well as the increasing popularity of ShapeNet (about 51,300 models) and Princeton ModelNet (127,915 models) in computer vision research. MedShapeNet is created as an alternative to these commonly used shape benchmarks to facilitate the translation of data-driven vision algorithms to medical applications, and it extends the opportunities to adapt SOTA vision algorithms to solve critical medical problems. Besides, the majority of the medical shapes in MedShapeNet are modeled directly on the imaging data of real patients, and therefore it complements well existing shape benchmarks comprising of computer-aided design (CAD) models. MedShapeNet currently includes more than 100,000 medical shapes, and provides annotations in the form of paired data. It is therefore also a freely available repository of 3D models for extended reality (virtual reality - VR, augmented reality - AR, mixed reality - MR) and medical 3D printing. This white paper describes in detail the motivations behind MedShapeNet, the shape acquisition procedures, the use cases, as well as the usage of the online shape search portal: https://medshapenet.ikim.nrw/
Open-Source Skull Reconstruction with MONAI
Nov 25, 2022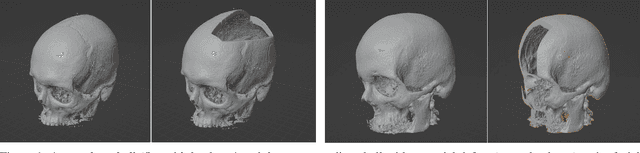
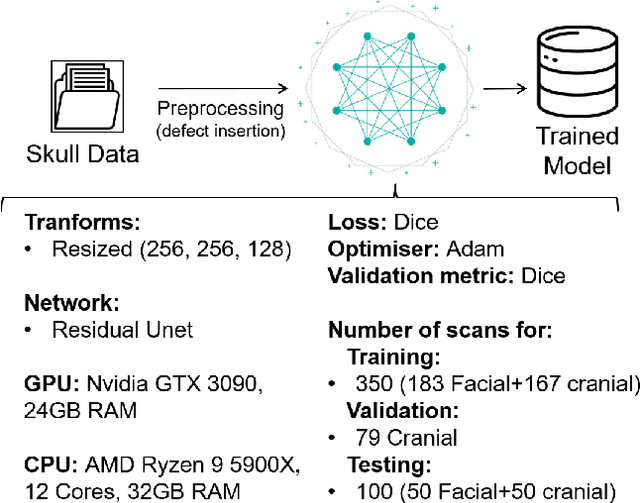
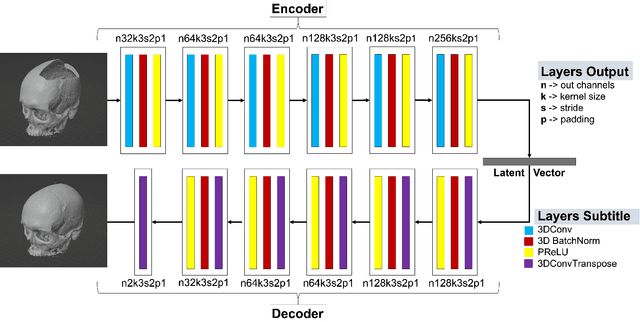
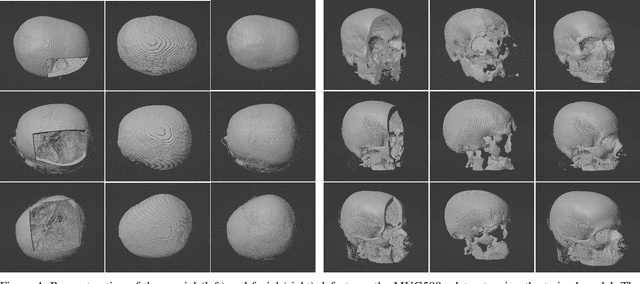
Abstract:We present a deep learning-based approach for skull reconstruction for MONAI, which has been pre-trained on the MUG500+ skull dataset. The implementation follows the MONAI contribution guidelines, hence, it can be easily tried out and used, and extended by MONAI users. The primary goal of this paper lies in the investigation of open-sourcing codes and pre-trained deep learning models under the MONAI framework. Nowadays, open-sourcing software, especially (pre-trained) deep learning models, has become increasingly important. Over the years, medical image analysis experienced a tremendous transformation. Over a decade ago, algorithms had to be implemented and optimized with low-level programming languages, like C or C++, to run in a reasonable time on a desktop PC, which was not as powerful as today's computers. Nowadays, users have high-level scripting languages like Python, and frameworks like PyTorch and TensorFlow, along with a sea of public code repositories at hand. As a result, implementations that had thousands of lines of C or C++ code in the past, can now be scripted with a few lines and in addition executed in a fraction of the time. To put this even on a higher level, the Medical Open Network for Artificial Intelligence (MONAI) framework tailors medical imaging research to an even more convenient process, which can boost and push the whole field. The MONAI framework is a freely available, community-supported, open-source and PyTorch-based framework, that also enables to provide research contributions with pre-trained models to others. Codes and pre-trained weights for skull reconstruction are publicly available at: https://github.com/Project-MONAI/research-contributions/tree/master/SkullRec
MOMO -- Deep Learning-driven classification of external DICOM studies for PACS archivation
Dec 01, 2021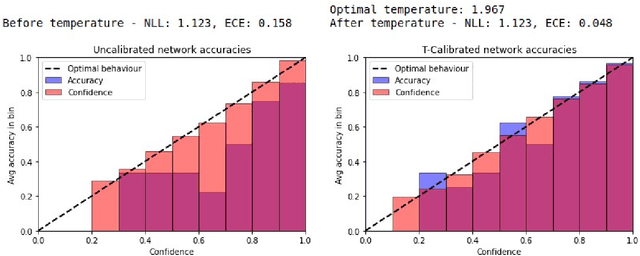
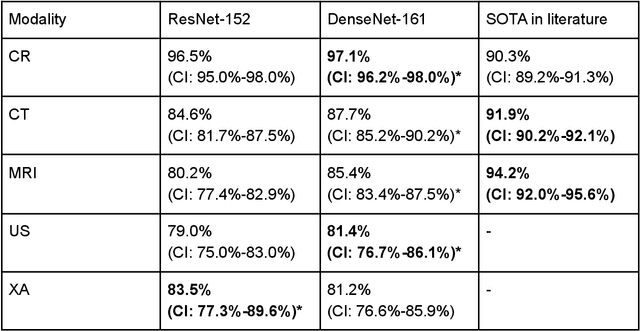
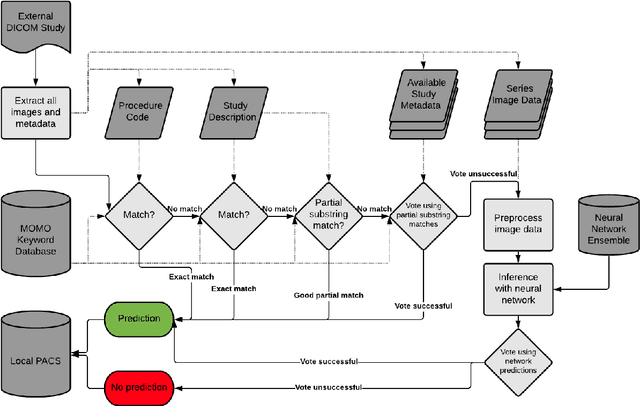
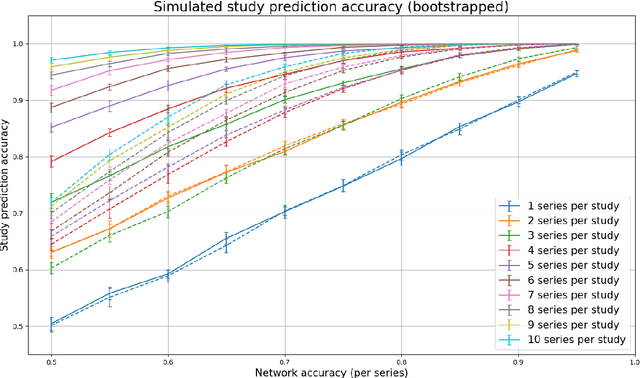
Abstract:Patients regularly continue assessment or treatment in other facilities than they began them in, receiving their previous imaging studies as a CD-ROM and requiring clinical staff at the new hospital to import these studies into their local database. However, between different facilities, standards for nomenclature, contents, or even medical procedures may vary, often requiring human intervention to accurately classify the received studies in the context of the recipient hospital's standards. In this study, the authors present MOMO (MOdality Mapping and Orchestration), a deep learning-based approach to automate this mapping process utilizing metadata substring matching and a neural network ensemble, which is trained to recognize the 76 most common imaging studies across seven different modalities. A retrospective study is performed to measure the accuracy that this algorithm can provide. To this end, a set of 11,934 imaging series with existing labels was retrieved from the local hospital's PACS database to train the neural networks. A set of 843 completely anonymized external studies was hand-labeled to assess the performance of our algorithm. Additionally, an ablation study was performed to measure the performance impact of the network ensemble in the algorithm, and a comparative performance test with a commercial product was conducted. In comparison to a commercial product (96.20% predictive power, 82.86% accuracy, 1.36% minor errors), a neural network ensemble alone performs the classification task with less accuracy (99.05% predictive power, 72.69% accuracy, 10.3% minor errors). However, MOMO outperforms either by a large margin in accuracy and with increased predictive power (99.29% predictive power, 92.71% accuracy, 2.63% minor errors).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge