Amr Abourayya
Protecting Sensitive Data through Federated Co-Training
Oct 09, 2023



Abstract:In many critical applications, sensitive data is inherently distributed. Federated learning trains a model collaboratively by aggregating the parameters of locally trained models. This avoids exposing sensitive local data. It is possible, though, to infer upon the sensitive data from the shared model parameters. At the same time, many types of machine learning models do not lend themselves to parameter aggregation, such as decision trees, or rule ensembles. It has been observed that in many applications, in particular healthcare, large unlabeled datasets are publicly available. They can be used to exchange information between clients by distributed distillation, i.e., co-regularizing local training via the discrepancy between the soft predictions of each local client on the unlabeled dataset. This, however, still discloses private information and restricts the types of models to those trainable via gradient-based methods. We propose to go one step further and use a form of federated co-training, where local hard labels on the public unlabeled datasets are shared and aggregated into a consensus label. This consensus label can be used for local training by any supervised machine learning model. We show that this federated co-training approach achieves a model quality comparable to both federated learning and distributed distillation on a set of benchmark datasets and real-world medical datasets. It improves privacy over both approaches, protecting against common membership inference attacks to the highest degree. Furthermore, we show that federated co-training can collaboratively train interpretable models, such as decision trees and rule ensembles, achieving a model quality comparable to centralized training.
MedShapeNet -- A Large-Scale Dataset of 3D Medical Shapes for Computer Vision
Sep 12, 2023



Abstract:We present MedShapeNet, a large collection of anatomical shapes (e.g., bones, organs, vessels) and 3D surgical instrument models. Prior to the deep learning era, the broad application of statistical shape models (SSMs) in medical image analysis is evidence that shapes have been commonly used to describe medical data. Nowadays, however, state-of-the-art (SOTA) deep learning algorithms in medical imaging are predominantly voxel-based. In computer vision, on the contrary, shapes (including, voxel occupancy grids, meshes, point clouds and implicit surface models) are preferred data representations in 3D, as seen from the numerous shape-related publications in premier vision conferences, such as the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), as well as the increasing popularity of ShapeNet (about 51,300 models) and Princeton ModelNet (127,915 models) in computer vision research. MedShapeNet is created as an alternative to these commonly used shape benchmarks to facilitate the translation of data-driven vision algorithms to medical applications, and it extends the opportunities to adapt SOTA vision algorithms to solve critical medical problems. Besides, the majority of the medical shapes in MedShapeNet are modeled directly on the imaging data of real patients, and therefore it complements well existing shape benchmarks comprising of computer-aided design (CAD) models. MedShapeNet currently includes more than 100,000 medical shapes, and provides annotations in the form of paired data. It is therefore also a freely available repository of 3D models for extended reality (virtual reality - VR, augmented reality - AR, mixed reality - MR) and medical 3D printing. This white paper describes in detail the motivations behind MedShapeNet, the shape acquisition procedures, the use cases, as well as the usage of the online shape search portal: https://medshapenet.ikim.nrw/
FAM: Relative Flatness Aware Minimization
Jul 05, 2023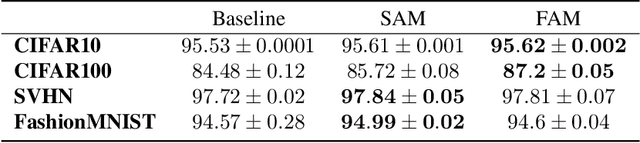
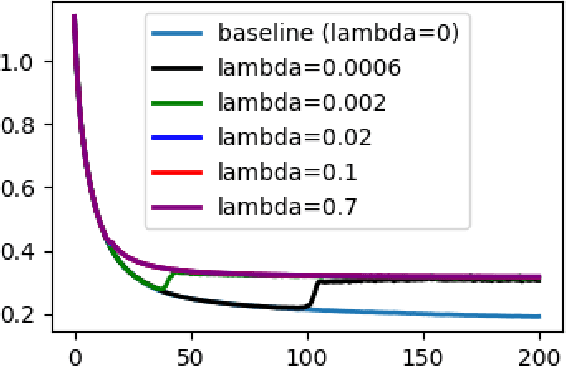
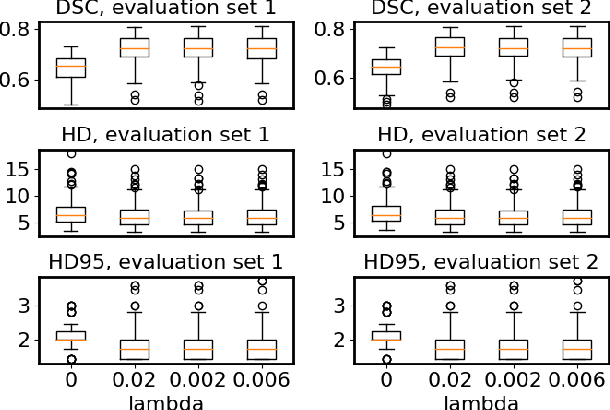
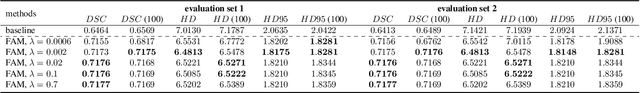
Abstract:Flatness of the loss curve around a model at hand has been shown to empirically correlate with its generalization ability. Optimizing for flatness has been proposed as early as 1994 by Hochreiter and Schmidthuber, and was followed by more recent successful sharpness-aware optimization techniques. Their widespread adoption in practice, though, is dubious because of the lack of theoretically grounded connection between flatness and generalization, in particular in light of the reparameterization curse - certain reparameterizations of a neural network change most flatness measures but do not change generalization. Recent theoretical work suggests that a particular relative flatness measure can be connected to generalization and solves the reparameterization curse. In this paper, we derive a regularizer based on this relative flatness that is easy to compute, fast, efficient, and works with arbitrary loss functions. It requires computing the Hessian only of a single layer of the network, which makes it applicable to large neural networks, and with it avoids an expensive mapping of the loss surface in the vicinity of the model. In an extensive empirical evaluation we show that this relative flatness aware minimization (FAM) improves generalization in a multitude of applications and models, both in finetuning and standard training. We make the code available at github.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge