Kaleb E. Smith
NVIDIA
MeDiSumQA: Patient-Oriented Question-Answer Generation from Discharge Letters
Feb 05, 2025Abstract:While increasing patients' access to medical documents improves medical care, this benefit is limited by varying health literacy levels and complex medical terminology. Large language models (LLMs) offer solutions by simplifying medical information. However, evaluating LLMs for safe and patient-friendly text generation is difficult due to the lack of standardized evaluation resources. To fill this gap, we developed MeDiSumQA. MeDiSumQA is a dataset created from MIMIC-IV discharge summaries through an automated pipeline combining LLM-based question-answer generation with manual quality checks. We use this dataset to evaluate various LLMs on patient-oriented question-answering. Our findings reveal that general-purpose LLMs frequently surpass biomedical-adapted models, while automated metrics correlate with human judgment. By releasing MeDiSumQA on PhysioNet, we aim to advance the development of LLMs to enhance patient understanding and ultimately improve care outcomes.
Comprehensive Study on German Language Models for Clinical and Biomedical Text Understanding
Apr 08, 2024



Abstract:Recent advances in natural language processing (NLP) can be largely attributed to the advent of pre-trained language models such as BERT and RoBERTa. While these models demonstrate remarkable performance on general datasets, they can struggle in specialized domains such as medicine, where unique domain-specific terminologies, domain-specific abbreviations, and varying document structures are common. This paper explores strategies for adapting these models to domain-specific requirements, primarily through continuous pre-training on domain-specific data. We pre-trained several German medical language models on 2.4B tokens derived from translated public English medical data and 3B tokens of German clinical data. The resulting models were evaluated on various German downstream tasks, including named entity recognition (NER), multi-label classification, and extractive question answering. Our results suggest that models augmented by clinical and translation-based pre-training typically outperform general domain models in medical contexts. We conclude that continuous pre-training has demonstrated the ability to match or even exceed the performance of clinical models trained from scratch. Furthermore, pre-training on clinical data or leveraging translated texts have proven to be reliable methods for domain adaptation in medical NLP tasks.
Machine Learning Methods Applied to Cortico-Cortical Evoked Potentials Aid in Localizing Seizure Onset Zones
Nov 15, 2022Abstract:Epilepsy affects millions of people, reducing quality of life and increasing risk of premature death. One-third of epilepsy cases are drug-resistant and require surgery for treatment, which necessitates localizing the seizure onset zone (SOZ) in the brain. Attempts have been made to use cortico-cortical evoked potentials (CCEPs) to improve SOZ localization but none have been successful enough for clinical adoption. Here, we compare the performance of ten machine learning classifiers in localizing SOZ from CCEP data. This preliminary study validates a novel application of machine learning, and the results establish our approach as a promising line of research that warrants further investigation. This work also serves to facilitate discussion and collaboration with fellow machine learning and/or epilepsy researchers.
Building an AI-ready RSE Workforce
Nov 09, 2021Abstract:Artificial Intelligence has been transforming industries and academic research across the globe, and research software development is no exception. Machine learning and deep learning are being applied in every aspect of the research software development lifecycles, from new algorithm design paradigms to software development processes. In this paper, we discuss our views on today's challenges and opportunities that AI has presented on research software development and engineers, and the approaches we, at the University of Florida, are taking to prepare our workforce for the new era of AI.
A Spectral Enabled GAN for Time Series Data Generation
Mar 02, 2021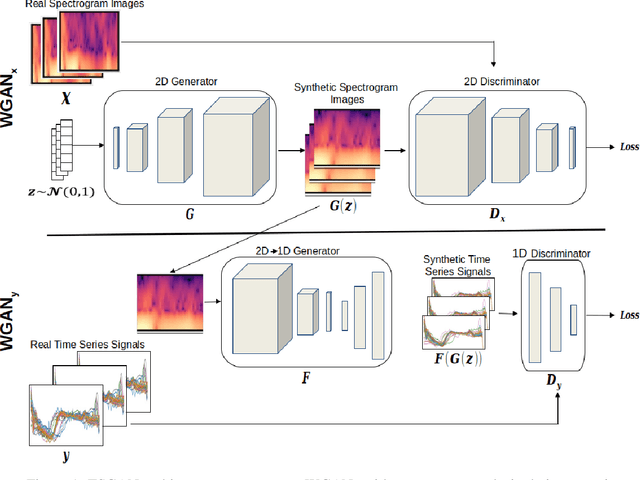
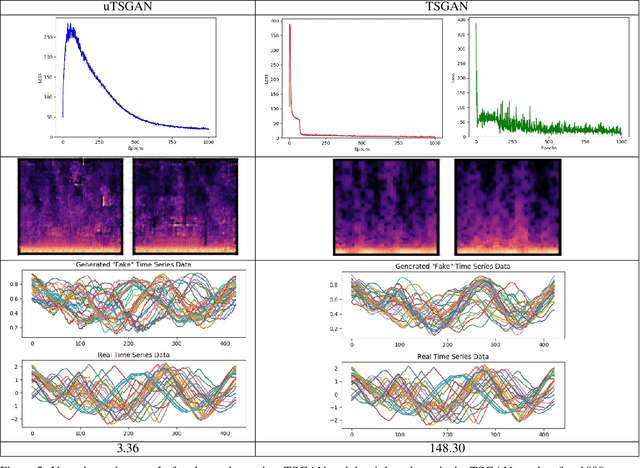
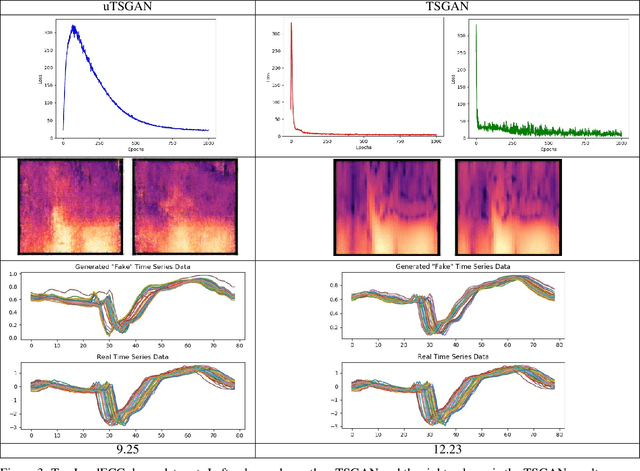
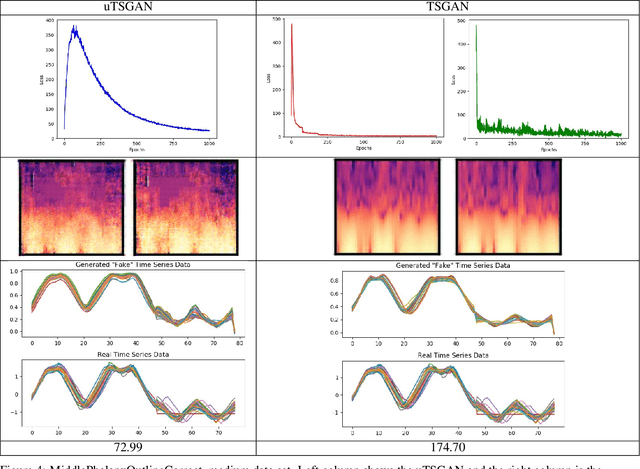
Abstract:Time dependent data is a main source of information in today's data driven world. Generating this type of data though has shown its challenges and made it an interesting research area in the field of generative machine learning. One such approach was that by Smith et al. who developed Time Series Generative Adversarial Network (TSGAN) which showed promising performance in generating time dependent data and the ability of few shot generation though being flawed in certain aspects of training and learning. This paper looks to improve on the results from TSGAN and address those flaws by unifying the training of the independent networks in TSGAN and creating a dependency both in training and learning. This improvement, called unified TSGAN (uTSGAN) was tested and comapred both quantitatively and qualitatively to its predecessor on 70 benchmark time series data sets used in the community. uTSGAN showed to outperform TSGAN in 80\% of the data sets by the same number of training epochs and 60\% of the data sets in 3/4th the amount of training time or less while maintaining the few shot generation ability with better FID scores across those data sets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge