Luise Modersohn
Deep learning and abstractive summarisation for radiological reports: an empirical study for adapting the PEGASUS models' family with scarce data
Sep 18, 2025Abstract:Regardless of the rapid development of artificial intelligence, abstractive summarisation is still challenging for sensitive and data-restrictive domains like medicine. With the increasing number of imaging, the relevance of automated tools for complex medical text summarisation is expected to become highly relevant. In this paper, we investigated the adaptation via fine-tuning process of a non-domain-specific abstractive summarisation encoder-decoder model family, and gave insights to practitioners on how to avoid over- and underfitting. We used PEGASUS and PEGASUS-X, on a medium-sized radiological reports public dataset. For each model, we comprehensively evaluated two different checkpoints with varying sizes of the same training data. We monitored the models' performances with lexical and semantic metrics during the training history on the fixed-size validation set. PEGASUS exhibited different phases, which can be related to epoch-wise double-descent, or peak-drop-recovery behaviour. For PEGASUS-X, we found that using a larger checkpoint led to a performance detriment. This work highlights the challenges and risks of fine-tuning models with high expressivity when dealing with scarce training data, and lays the groundwork for future investigations into more robust fine-tuning strategies for summarisation models in specialised domains.
Large Language Models-Enabled Digital Twins for Precision Medicine in Rare Gynecological Tumors
Aug 31, 2024Abstract:Rare gynecological tumors (RGTs) present major clinical challenges due to their low incidence and heterogeneity. The lack of clear guidelines leads to suboptimal management and poor prognosis. Molecular tumor boards accelerate access to effective therapies by tailoring treatment based on biomarkers, beyond cancer type. Unstructured data that requires manual curation hinders efficient use of biomarker profiling for therapy matching. This study explores the use of large language models (LLMs) to construct digital twins for precision medicine in RGTs. Our proof-of-concept digital twin system integrates clinical and biomarker data from institutional and published cases (n=21) and literature-derived data (n=655 publications with n=404,265 patients) to create tailored treatment plans for metastatic uterine carcinosarcoma, identifying options potentially missed by traditional, single-source analysis. LLM-enabled digital twins efficiently model individual patient trajectories. Shifting to a biology-based rather than organ-based tumor definition enables personalized care that could advance RGT management and thus enhance patient outcomes.
Towards a Unified Framework for Emotion Analysis
Dec 01, 2020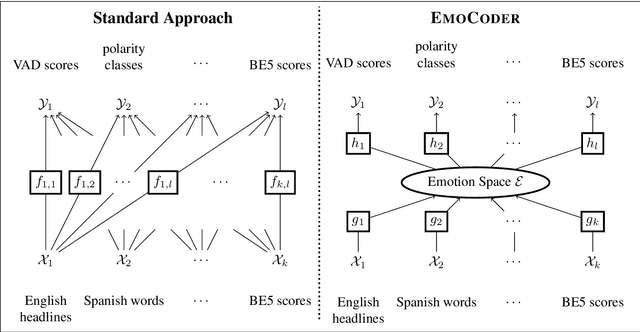
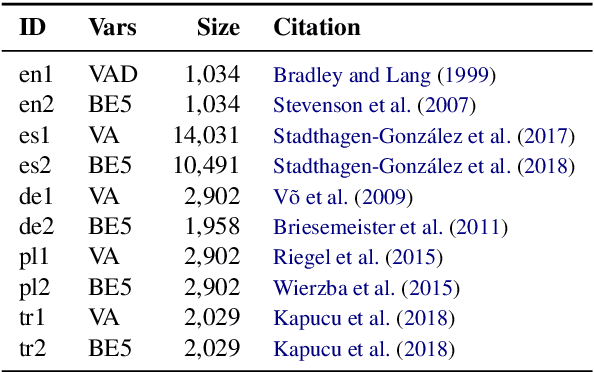
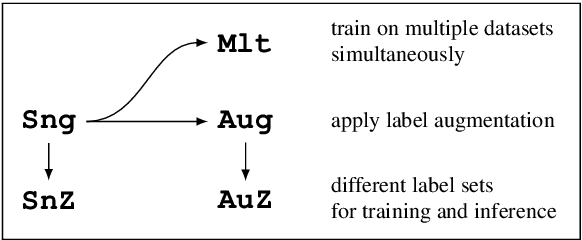
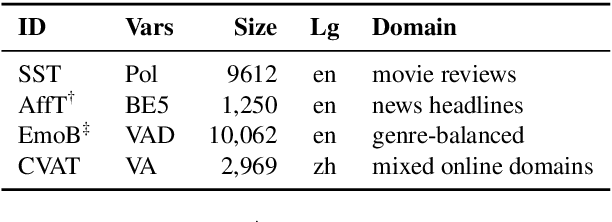
Abstract:We present EmoCoder, a modular encoder-decoder architecture that generalizes emotion analysis over different tasks (sentence-level, word-level, label-to-label mapping), domains (natural languages and their registers), and label formats (e.g., polarity classes, basic emotions, and affective dimensions). Experiments on 14 datasets indicate that EmoCoder learns an interpretable language-independent representation of emotions, allows seamless absorption of state-of-the-art models, and maintains strong prediction quality, even when tested on unseen combinations of domains and label formats.
GGPONC: A Corpus of German Medical Text with Rich Metadata Based on Clinical Practice Guidelines
Jul 13, 2020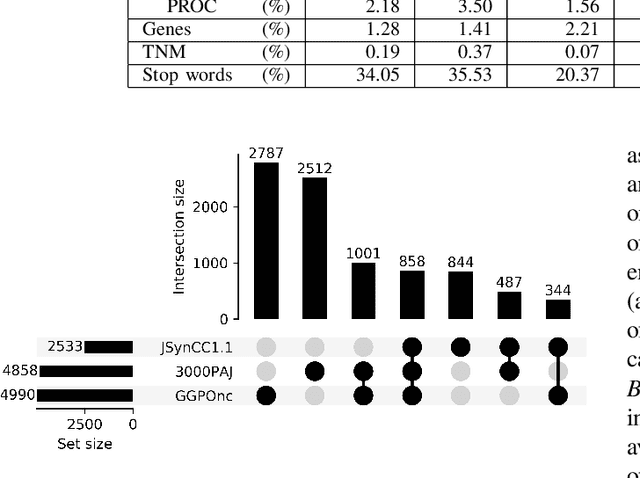
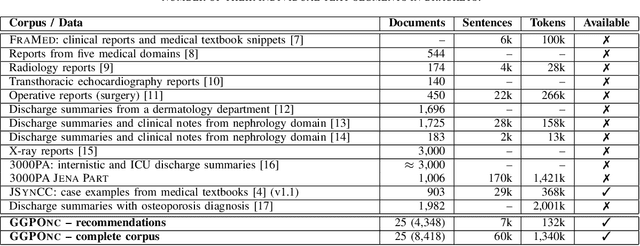
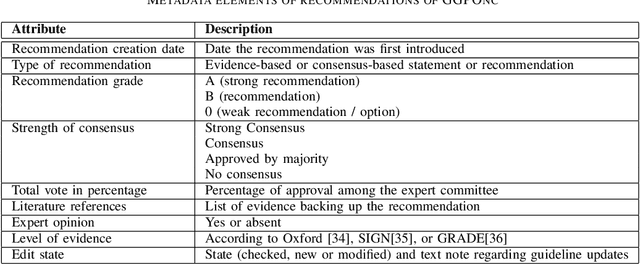
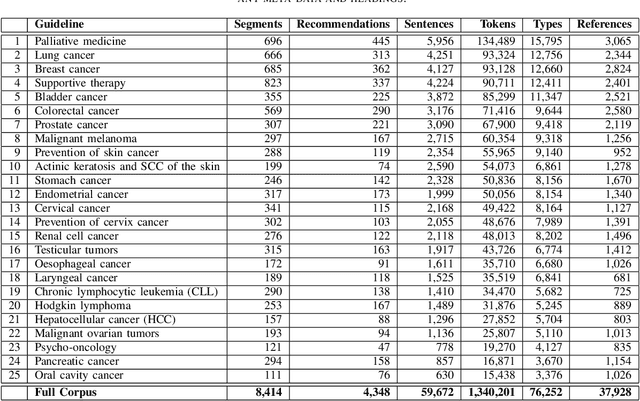
Abstract:The lack of publicly available text corpora is a major obstacle for progress in clinical natural language processing, for non-English speaking countries in particular. In this work, we present GGPONC (German Guideline Program in Oncology NLP Corpus), a freely distributable German language corpus based on clinical practice guidelines in the field of oncology. The corpus is one of the largest corpora of German medical text to date. It does not contain any patient-related data and can therefore be used without data protection restrictions. Moreover, it is the first corpus for the German language covering diverse conditions in a large medical subfield. In addition to the textual sources, we provide a large variety of metadata, such as literature references and evidence levels. By applying and evaluating existing medical information extraction pipelines for German text, we are able to draw comparisons for the use of medical language to other medical text corpora.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge