Matthieu-P. Schapranow
xMEN: A Modular Toolkit for Cross-Lingual Medical Entity Normalization
Oct 17, 2023Abstract:Objective: To improve performance of medical entity normalization across many languages, especially when fewer language resources are available compared to English. Materials and Methods: We introduce xMEN, a modular system for cross-lingual medical entity normalization, which performs well in both low- and high-resource scenarios. When synonyms in the target language are scarce for a given terminology, we leverage English aliases via cross-lingual candidate generation. For candidate ranking, we incorporate a trainable cross-encoder model if annotations for the target task are available. We also evaluate cross-encoders trained in a weakly supervised manner based on machine-translated datasets from a high resource domain. Our system is publicly available as an extensible Python toolkit. Results: xMEN improves the state-of-the-art performance across a wide range of multilingual benchmark datasets. Weakly supervised cross-encoders are effective when no training data is available for the target task. Through the compatibility of xMEN with the BigBIO framework, it can be easily used with existing and prospective datasets. Discussion: Our experiments show the importance of balancing the output of general-purpose candidate generators with subsequent trainable re-rankers, which we achieve through a rank regularization term in the loss function of the cross-encoder. However, error analysis reveals that multi-word expressions and other complex entities are still challenging. Conclusion: xMEN exhibits strong performance for medical entity normalization in multiple languages, even when no labeled data and few terminology aliases for the target language are available. Its configuration system and evaluation modules enable reproducible benchmarks. Models and code are available online at the following URL: https://github.com/hpi-dhc/xmen
GGPONC: A Corpus of German Medical Text with Rich Metadata Based on Clinical Practice Guidelines
Jul 13, 2020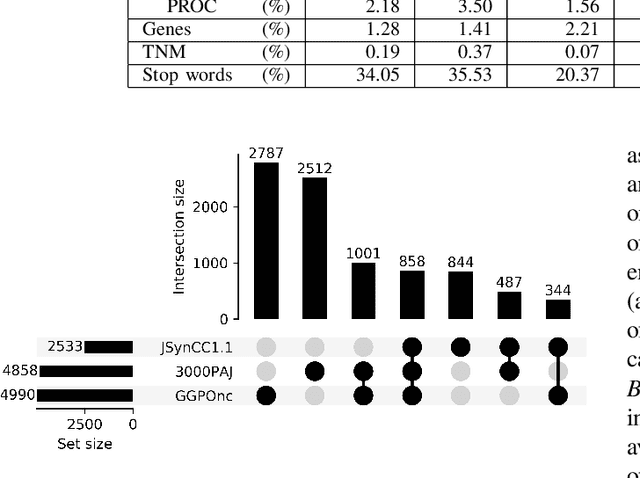
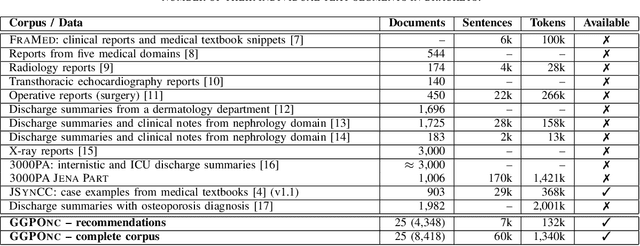
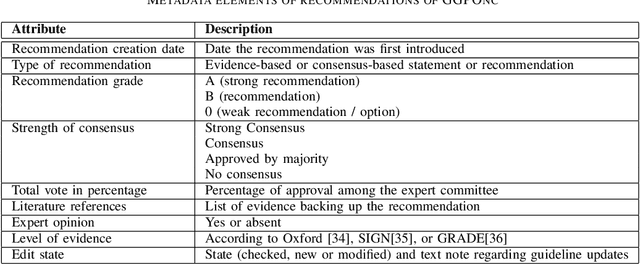
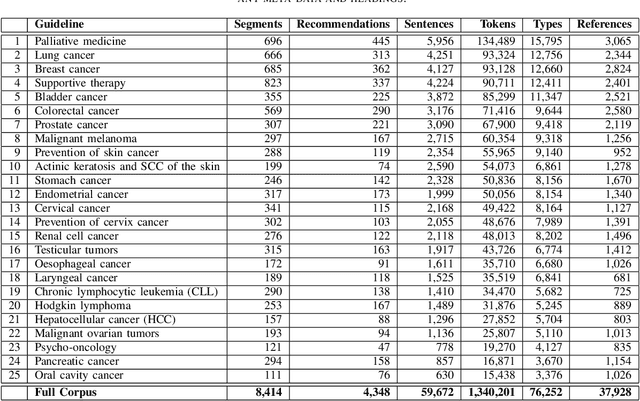
Abstract:The lack of publicly available text corpora is a major obstacle for progress in clinical natural language processing, for non-English speaking countries in particular. In this work, we present GGPONC (German Guideline Program in Oncology NLP Corpus), a freely distributable German language corpus based on clinical practice guidelines in the field of oncology. The corpus is one of the largest corpora of German medical text to date. It does not contain any patient-related data and can therefore be used without data protection restrictions. Moreover, it is the first corpus for the German language covering diverse conditions in a large medical subfield. In addition to the textual sources, we provide a large variety of metadata, such as literature references and evidence levels. By applying and evaluating existing medical information extraction pipelines for German text, we are able to draw comparisons for the use of medical language to other medical text corpora.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge