Bert Arnrich
German Text Embedding Clustering Benchmark
Jan 05, 2024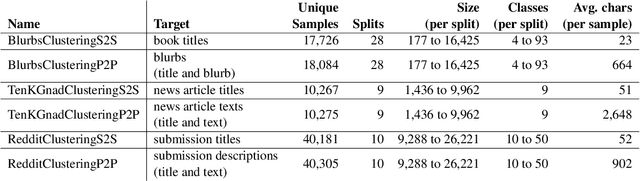
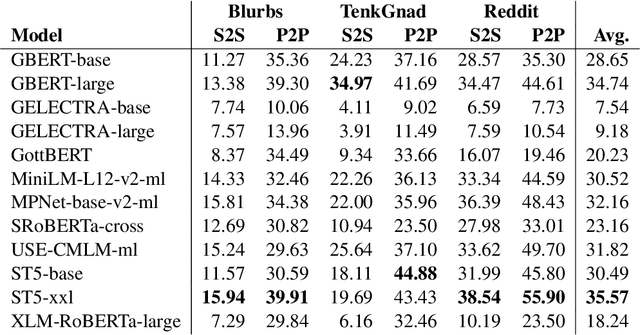
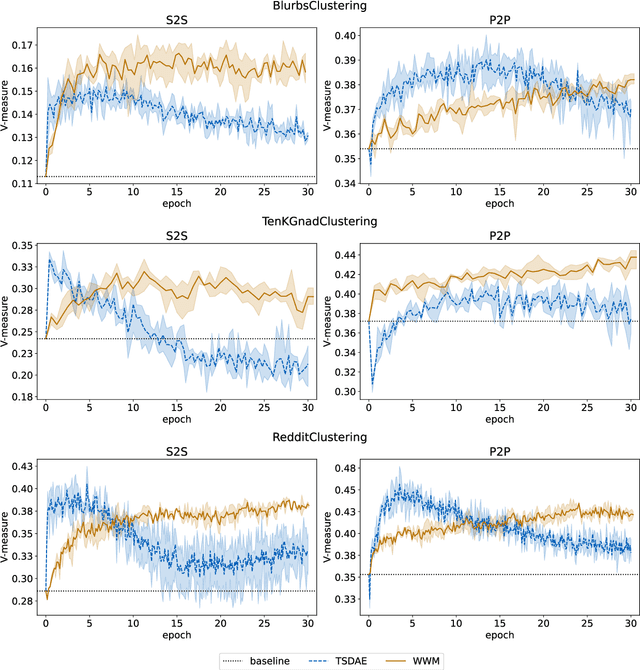
Abstract:This work introduces a benchmark assessing the performance of clustering German text embeddings in different domains. This benchmark is driven by the increasing use of clustering neural text embeddings in tasks that require the grouping of texts (such as topic modeling) and the need for German resources in existing benchmarks. We provide an initial analysis for a range of pre-trained mono- and multilingual models evaluated on the outcome of different clustering algorithms. Results include strong performing mono- and multilingual models. Reducing the dimensions of embeddings can further improve clustering. Additionally, we conduct experiments with continued pre-training for German BERT models to estimate the benefits of this additional training. Our experiments suggest that significant performance improvements are possible for short text. All code and datasets are publicly available.
xMEN: A Modular Toolkit for Cross-Lingual Medical Entity Normalization
Oct 17, 2023Abstract:Objective: To improve performance of medical entity normalization across many languages, especially when fewer language resources are available compared to English. Materials and Methods: We introduce xMEN, a modular system for cross-lingual medical entity normalization, which performs well in both low- and high-resource scenarios. When synonyms in the target language are scarce for a given terminology, we leverage English aliases via cross-lingual candidate generation. For candidate ranking, we incorporate a trainable cross-encoder model if annotations for the target task are available. We also evaluate cross-encoders trained in a weakly supervised manner based on machine-translated datasets from a high resource domain. Our system is publicly available as an extensible Python toolkit. Results: xMEN improves the state-of-the-art performance across a wide range of multilingual benchmark datasets. Weakly supervised cross-encoders are effective when no training data is available for the target task. Through the compatibility of xMEN with the BigBIO framework, it can be easily used with existing and prospective datasets. Discussion: Our experiments show the importance of balancing the output of general-purpose candidate generators with subsequent trainable re-rankers, which we achieve through a rank regularization term in the loss function of the cross-encoder. However, error analysis reveals that multi-word expressions and other complex entities are still challenging. Conclusion: xMEN exhibits strong performance for medical entity normalization in multiple languages, even when no labeled data and few terminology aliases for the target language are available. Its configuration system and evaluation modules enable reproducible benchmarks. Models and code are available online at the following URL: https://github.com/hpi-dhc/xmen
A Real-time Human Pose Estimation Approach for Optimal Sensor Placement in Sensor-based Human Activity Recognition
Jul 06, 2023Abstract:Sensor-based Human Activity Recognition facilitates unobtrusive monitoring of human movements. However, determining the most effective sensor placement for optimal classification performance remains challenging. This paper introduces a novel methodology to resolve this issue, using real-time 2D pose estimations derived from video recordings of target activities. The derived skeleton data provides a unique strategy for identifying the optimal sensor location. We validate our approach through a feasibility study, applying inertial sensors to monitor 13 different activities across ten subjects. Our findings indicate that the vision-based method for sensor placement offers comparable results to the conventional deep learning approach, demonstrating its efficacy. This research significantly advances the field of Human Activity Recognition by providing a lightweight, on-device solution for determining the optimal sensor placement, thereby enhancing data anonymization and supporting a multimodal classification approach.
Yet Another ICU Benchmark: A Flexible Multi-Center Framework for Clinical ML
Jun 08, 2023Abstract:Medical applications of machine learning (ML) have experienced a surge in popularity in recent years. The intensive care unit (ICU) is a natural habitat for ML given the abundance of available data from electronic health records. Models have been proposed to address numerous ICU prediction tasks like the early detection of complications. While authors frequently report state-of-the-art performance, it is challenging to verify claims of superiority. Datasets and code are not always published, and cohort definitions, preprocessing pipelines, and training setups are difficult to reproduce. This work introduces Yet Another ICU Benchmark (YAIB), a modular framework that allows researchers to define reproducible and comparable clinical ML experiments; we offer an end-to-end solution from cohort definition to model evaluation. The framework natively supports most open-access ICU datasets (MIMIC III/IV, eICU, HiRID, AUMCdb) and is easily adaptable to future ICU datasets. Combined with a transparent preprocessing pipeline and extensible training code for multiple ML and deep learning models, YAIB enables unified model development. Our benchmark comes with five predefined established prediction tasks (mortality, acute kidney injury, sepsis, kidney function, and length of stay) developed in collaboration with clinicians. Adding further tasks is straightforward by design. Using YAIB, we demonstrate that the choice of dataset, cohort definition, and preprocessing have a major impact on the prediction performance - often more so than model class - indicating an urgent need for YAIB as a holistic benchmarking tool. We provide our work to the clinical ML community to accelerate method development and enable real-world clinical implementations. Software Repository: https://github.com/rvandewater/YAIB.
DPD-fVAE: Synthetic Data Generation Using Federated Variational Autoencoders With Differentially-Private Decoder
Nov 21, 2022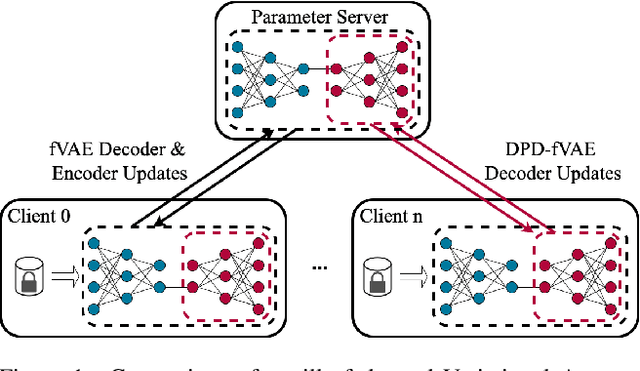

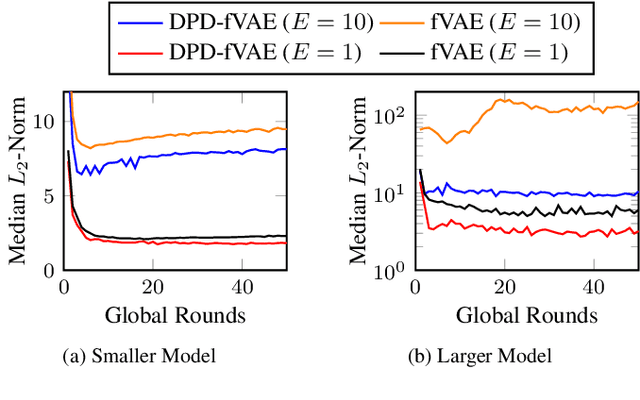
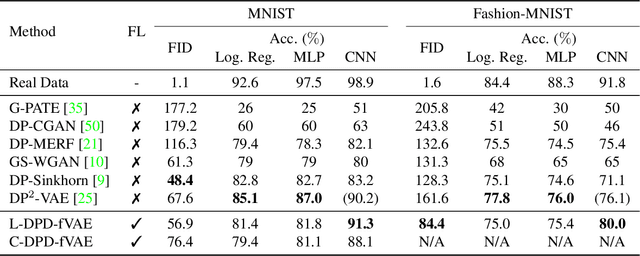
Abstract:Federated learning (FL) is getting increased attention for processing sensitive, distributed datasets common to domains such as healthcare. Instead of directly training classification models on these datasets, recent works have considered training data generators capable of synthesising a new dataset which is not protected by any privacy restrictions. Thus, the synthetic data can be made available to anyone, which enables further evaluation of machine learning architectures and research questions off-site. As an additional layer of privacy-preservation, differential privacy can be introduced into the training process. We propose DPD-fVAE, a federated Variational Autoencoder with Differentially-Private Decoder, to synthesise a new, labelled dataset for subsequent machine learning tasks. By synchronising only the decoder component with FL, we can reduce the privacy cost per epoch and thus enable better data generators. In our evaluation on MNIST, Fashion-MNIST and CelebA, we show the benefits of DPD-fVAE and report competitive performance to related work in terms of Fr\'echet Inception Distance and accuracy of classifiers trained on the synthesised dataset.
Defending against Reconstruction Attacks through Differentially Private Federated Learning for Classification of Heterogeneous Chest X-Ray Data
May 06, 2022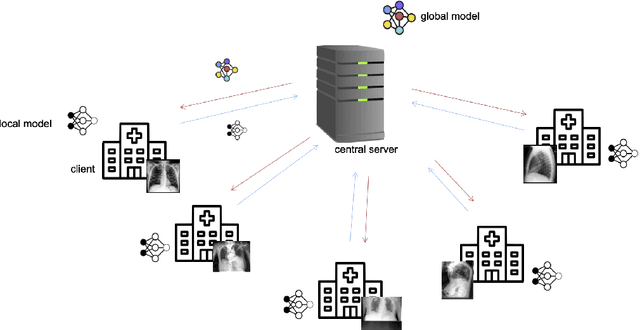
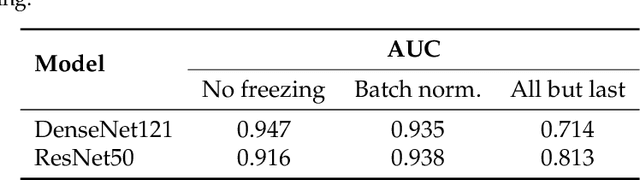
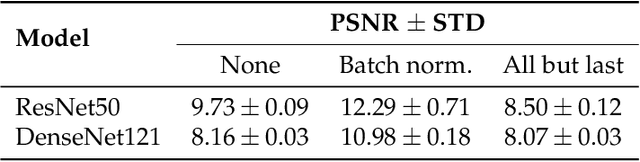
Abstract:Privacy regulations and the physical distribution of heterogeneous data are often primary concerns for the development of deep learning models in a medical context. This paper evaluates the feasibility of differentially private federated learning for chest X-ray classification as a defense against privacy attacks on DenseNet121 and ResNet50 network architectures. We simulated a federated environment by distributing images from the public CheXpert and Mendeley chest X-ray datasets unevenly among 36 clients. Both non-private baseline models achieved an area under the ROC curve (AUC) of 0.94 on the binary classification task of detecting the presence of a medical finding. We demonstrate that both model architectures are vulnerable to privacy violation by applying image reconstruction attacks to local model updates from individual clients. The attack was particularly successful during later training stages. To mitigate the risk of privacy breach, we integrated R\'enyi differential privacy with a Gaussian noise mechanism into local model training. We evaluate model performance and attack vulnerability for privacy budgets $\epsilon \in$ {1, 3, 6, 10}. The DenseNet121 achieved the best utility-privacy trade-off with an AUC of 0.94 for $\epsilon$ = 6. Model performance deteriorated slightly for individual clients compared to the non-private baseline. The ResNet50 only reached an AUC of 0.76 in the same privacy setting. Its performance was inferior to that of the DenseNet121 for all considered privacy constraints, suggesting that the DenseNet121 architecture is more robust to differentially private training.
Implicit Model Specialization through DAG-based Decentralized Federated Learning
Nov 03, 2021
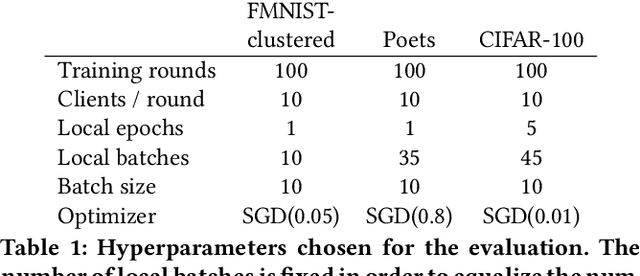
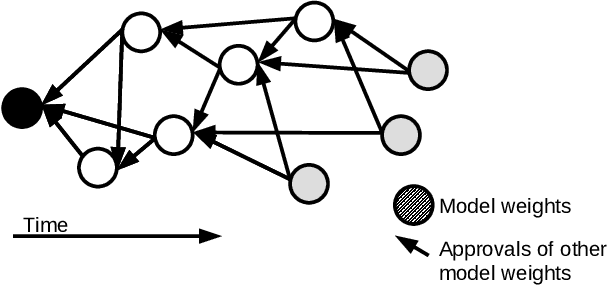

Abstract:Federated learning allows a group of distributed clients to train a common machine learning model on private data. The exchange of model updates is managed either by a central entity or in a decentralized way, e.g. by a blockchain. However, the strong generalization across all clients makes these approaches unsuited for non-independent and identically distributed (non-IID) data. We propose a unified approach to decentralization and personalization in federated learning that is based on a directed acyclic graph (DAG) of model updates. Instead of training a single global model, clients specialize on their local data while using the model updates from other clients dependent on the similarity of their respective data. This specialization implicitly emerges from the DAG-based communication and selection of model updates. Thus, we enable the evolution of specialized models, which focus on a subset of the data and therefore cover non-IID data better than federated learning in a centralized or blockchain-based setup. To the best of our knowledge, the proposed solution is the first to unite personalization and poisoning robustness in fully decentralized federated learning. Our evaluation shows that the specialization of models emerges directly from the DAG-based communication of model updates on three different datasets. Furthermore, we show stable model accuracy and less variance across clients when compared to federated averaging.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge