Roland Roller
Temporal Fusion Nexus: A task-agnostic multi-modal embedding model for clinical narratives and irregular time series in post-kidney transplant care
Jan 13, 2026Abstract:We introduce Temporal Fusion Nexus (TFN), a multi-modal and task-agnostic embedding model to integrate irregular time series and unstructured clinical narratives. We analysed TFN in post-kidney transplant (KTx) care, with a retrospective cohort of 3382 patients, on three key outcomes: graft loss, graft rejection, and mortality. Compared to state-of-the-art model in post KTx care, TFN achieved higher performance for graft loss (AUC 0.96 vs. 0.94) and graft rejection (AUC 0.84 vs. 0.74). In mortality prediction, TFN yielded an AUC of 0.86. TFN outperformed unimodal baselines (approx 10% AUC improvement over time series only baseline, approx 5% AUC improvement over time series with static patient data). Integrating clinical text improved performance across all tasks. Disentanglement metrics confirmed robust and interpretable latent factors in the embedding space, and SHAP-based attributions confirmed alignment with clinical reasoning. TFN has potential application in clinical tasks beyond KTx, where heterogeneous data sources, irregular longitudinal data, and rich narrative documentation are available.
Detecting Pipeline Failures through Fine-Grained Analysis of Web Agents
Sep 17, 2025Abstract:Web agents powered by large language models (LLMs) can autonomously perform complex, multistep tasks in dynamic web environments. However, current evaluations mostly focus on the overall success while overlooking intermediate errors. This limits insight into failure modes and hinders systematic improvement. This work analyzes existing benchmarks and highlights the lack of fine-grained diagnostic tools. To address this gap, we propose a modular evaluation framework that decomposes agent pipelines into interpretable stages for detailed error analysis. Using the SeeAct framework and the Mind2Web dataset as a case study, we show how this approach reveals actionable weaknesses missed by standard metrics - paving the way for more robust and generalizable web agents.
Infherno: End-to-end Agent-based FHIR Resource Synthesis from Free-form Clinical Notes
Jul 16, 2025Abstract:For clinical data integration and healthcare services, the HL7 FHIR standard has established itself as a desirable format for interoperability between complex health data. Previous attempts at automating the translation from free-form clinical notes into structured FHIR resources rely on modular, rule-based systems or LLMs with instruction tuning and constrained decoding. Since they frequently suffer from limited generalizability and structural inconformity, we propose an end-to-end framework powered by LLM agents, code execution, and healthcare terminology database tools to address these issues. Our solution, called Infherno, is designed to adhere to the FHIR document schema and competes well with a human baseline in predicting FHIR resources from unstructured text. The implementation features a front end for custom and synthetic data and both local and proprietary models, supporting clinical data integration processes and interoperability across institutions.
One Size Fits None: Rethinking Fairness in Medical AI
Jun 17, 2025Abstract:Machine learning (ML) models are increasingly used to support clinical decision-making. However, real-world medical datasets are often noisy, incomplete, and imbalanced, leading to performance disparities across patient subgroups. These differences raise fairness concerns, particularly when they reinforce existing disadvantages for marginalized groups. In this work, we analyze several medical prediction tasks and demonstrate how model performance varies with patient characteristics. While ML models may demonstrate good overall performance, we argue that subgroup-level evaluation is essential before integrating them into clinical workflows. By conducting a performance analysis at the subgroup level, differences can be clearly identified-allowing, on the one hand, for performance disparities to be considered in clinical practice, and on the other hand, for these insights to inform the responsible development of more effective models. Thereby, our work contributes to a practical discussion around the subgroup-sensitive development and deployment of medical ML models and the interconnectedness of fairness and transparency.
Beyond De-Identification: A Structured Approach for Defining and Detecting Indirect Identifiers in Medical Texts
Feb 18, 2025
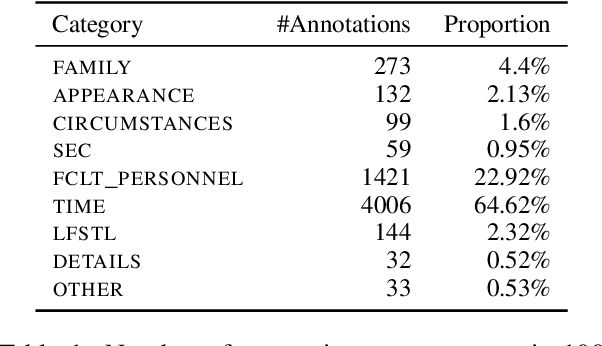
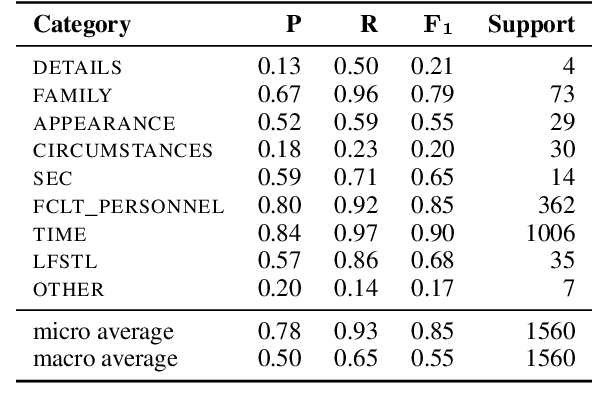

Abstract:Sharing sensitive texts for scientific purposes requires appropriate techniques to protect the privacy of patients and healthcare personnel. Anonymizing textual data is particularly challenging due to the presence of diverse unstructured direct and indirect identifiers. To mitigate the risk of re-identification, this work introduces a schema of nine categories of indirect identifiers designed to account for different potential adversaries, including acquaintances, family members and medical staff. Using this schema, we annotate 100 MIMIC-III discharge summaries and propose baseline models for identifying indirect identifiers. We will release the annotation guidelines, annotation spans (6,199 annotations in total) and the corresponding MIMIC-III document IDs to support further research in this area.
A Dataset for Pharmacovigilance in German, French, and Japanese: Annotating Adverse Drug Reactions across Languages
Mar 27, 2024



Abstract:User-generated data sources have gained significance in uncovering Adverse Drug Reactions (ADRs), with an increasing number of discussions occurring in the digital world. However, the existing clinical corpora predominantly revolve around scientific articles in English. This work presents a multilingual corpus of texts concerning ADRs gathered from diverse sources, including patient fora, social media, and clinical reports in German, French, and Japanese. Our corpus contains annotations covering 12 entity types, four attribute types, and 13 relation types. It contributes to the development of real-world multilingual language models for healthcare. We provide statistics to highlight certain challenges associated with the corpus and conduct preliminary experiments resulting in strong baselines for extracting entities and relations between these entities, both within and across languages.
xMEN: A Modular Toolkit for Cross-Lingual Medical Entity Normalization
Oct 17, 2023Abstract:Objective: To improve performance of medical entity normalization across many languages, especially when fewer language resources are available compared to English. Materials and Methods: We introduce xMEN, a modular system for cross-lingual medical entity normalization, which performs well in both low- and high-resource scenarios. When synonyms in the target language are scarce for a given terminology, we leverage English aliases via cross-lingual candidate generation. For candidate ranking, we incorporate a trainable cross-encoder model if annotations for the target task are available. We also evaluate cross-encoders trained in a weakly supervised manner based on machine-translated datasets from a high resource domain. Our system is publicly available as an extensible Python toolkit. Results: xMEN improves the state-of-the-art performance across a wide range of multilingual benchmark datasets. Weakly supervised cross-encoders are effective when no training data is available for the target task. Through the compatibility of xMEN with the BigBIO framework, it can be easily used with existing and prospective datasets. Discussion: Our experiments show the importance of balancing the output of general-purpose candidate generators with subsequent trainable re-rankers, which we achieve through a rank regularization term in the loss function of the cross-encoder. However, error analysis reveals that multi-word expressions and other complex entities are still challenging. Conclusion: xMEN exhibits strong performance for medical entity normalization in multiple languages, even when no labeled data and few terminology aliases for the target language are available. Its configuration system and evaluation modules enable reproducible benchmarks. Models and code are available online at the following URL: https://github.com/hpi-dhc/xmen
Factuality Detection using Machine Translation -- a Use Case for German Clinical Text
Aug 17, 2023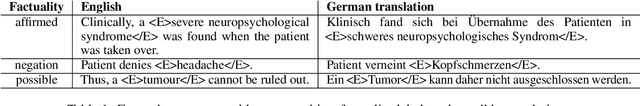
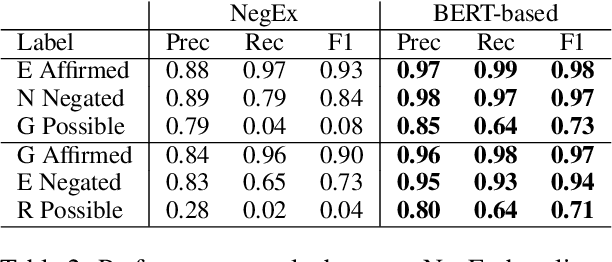
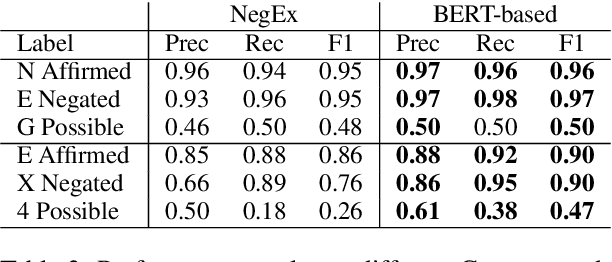
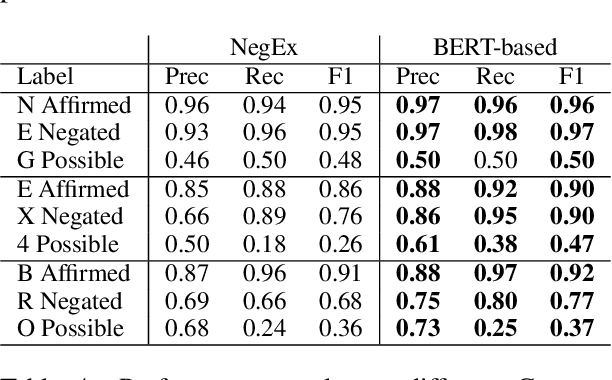
Abstract:Factuality can play an important role when automatically processing clinical text, as it makes a difference if particular symptoms are explicitly not present, possibly present, not mentioned, or affirmed. In most cases, a sufficient number of examples is necessary to handle such phenomena in a supervised machine learning setting. However, as clinical text might contain sensitive information, data cannot be easily shared. In the context of factuality detection, this work presents a simple solution using machine translation to translate English data to German to train a transformer-based factuality detection model.
Cross-lingual Approaches for the Detection of Adverse Drug Reactions in German from a Patient's Perspective
Aug 03, 2022
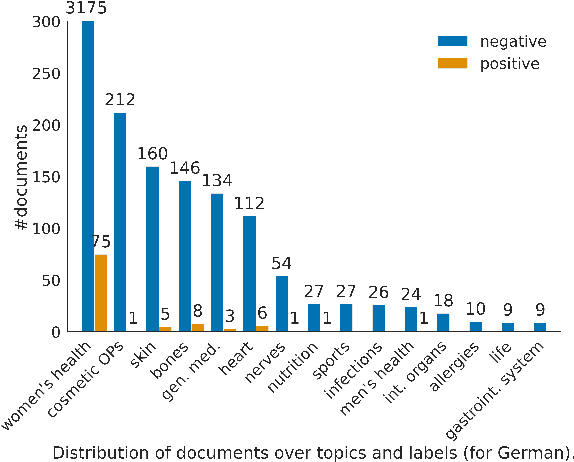
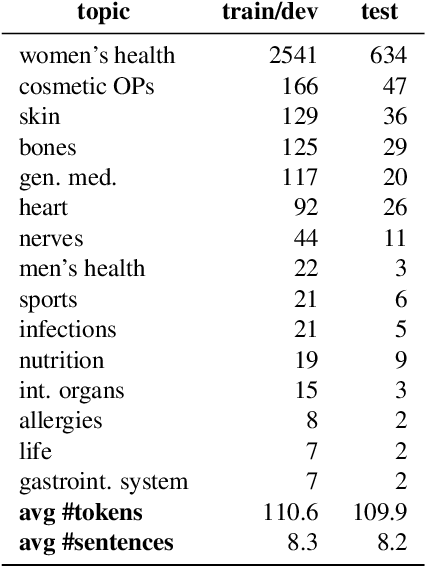
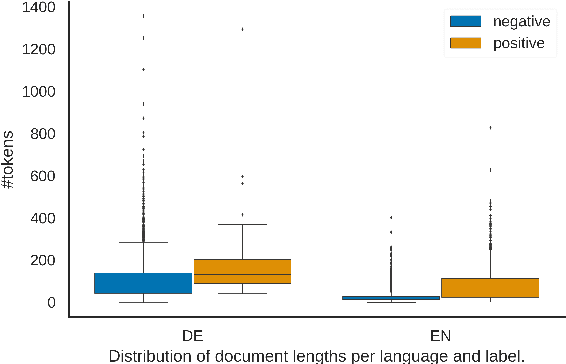
Abstract:In this work, we present the first corpus for German Adverse Drug Reaction (ADR) detection in patient-generated content. The data consists of 4,169 binary annotated documents from a German patient forum, where users talk about health issues and get advice from medical doctors. As is common in social media data in this domain, the class labels of the corpus are very imbalanced. This and a high topic imbalance make it a very challenging dataset, since often, the same symptom can have several causes and is not always related to a medication intake. We aim to encourage further multi-lingual efforts in the domain of ADR detection and provide preliminary experiments for binary classification using different methods of zero- and few-shot learning based on a multi-lingual model. When fine-tuning XLM-RoBERTa first on English patient forum data and then on the new German data, we achieve an F1-score of 37.52 for the positive class. We make the dataset and models publicly available for the community.
A Medical Information Extraction Workbench to Process German Clinical Text
Jul 08, 2022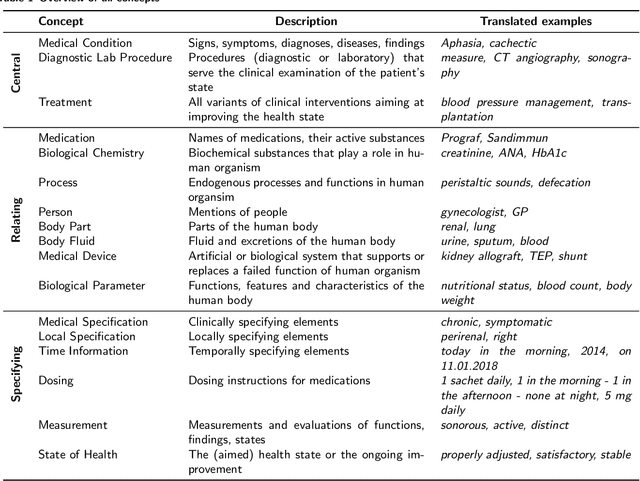
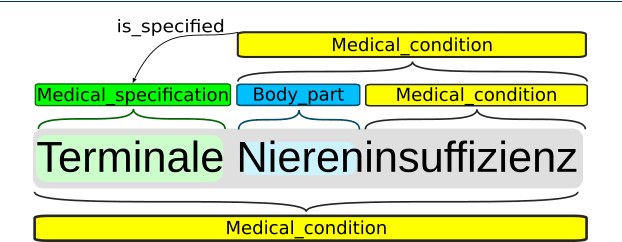

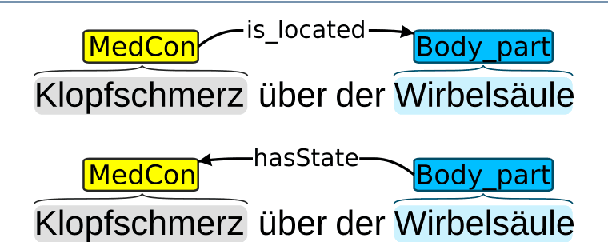
Abstract:Background: In the information extraction and natural language processing domain, accessible datasets are crucial to reproduce and compare results. Publicly available implementations and tools can serve as benchmark and facilitate the development of more complex applications. However, in the context of clinical text processing the number of accessible datasets is scarce -- and so is the number of existing tools. One of the main reasons is the sensitivity of the data. This problem is even more evident for non-English languages. Approach: In order to address this situation, we introduce a workbench: a collection of German clinical text processing models. The models are trained on a de-identified corpus of German nephrology reports. Result: The presented models provide promising results on in-domain data. Moreover, we show that our models can be also successfully applied to other biomedical text in German. Our workbench is made publicly available so it can be used out of the box, as a benchmark or transferred to related problems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge