Pierre Zweigenbaum
LISN
Kad: A Framework for Proxy-based Test-time Alignment with Knapsack Approximation Deferral
Oct 30, 2025Abstract:Several previous works concluded that the largest part of generation capabilities of large language models (LLM) are learned (early) during pre-training. However, LLMs still require further alignment to adhere to downstream task requirements and stylistic preferences, among other desired properties. As LLMs continue to scale in terms of size, the computational cost of alignment procedures increase prohibitively. In this work, we propose a novel approach to circumvent these costs via proxy-based test-time alignment, i.e. using guidance from a small aligned model. Our approach can be described as token-specific cascading method, where the token-specific deferral rule is reduced to 0-1 knapsack problem. In this setting, we derive primal and dual approximations of the optimal deferral decision. We experimentally show the benefits of our method both in task performance and speculative decoding speed.
Am I eligible? Natural Language Inference for Clinical Trial Patient Recruitment: the Patient's Point of View
Mar 19, 2025



Abstract:Recruiting patients to participate in clinical trials can be challenging and time-consuming. Usually, participation in a clinical trial is initiated by a healthcare professional and proposed to the patient. Promoting clinical trials directly to patients via online recruitment might help to reach them more efficiently. In this study, we address the case where a patient is initiating their own recruitment process and wants to determine whether they are eligible for a given clinical trial, using their own language to describe their medical profile. To study whether this creates difficulties in the patient trial matching process, we design a new dataset and task, Natural Language Inference for Patient Recruitment (NLI4PR), in which patient language profiles must be matched to clinical trials. We create it by adapting the TREC 2022 Clinical Trial Track dataset, which provides patients' medical profiles, and rephrasing them manually using patient language. We also use the associated clinical trial reports where the patients are either eligible or excluded. We prompt several open-source Large Language Models on our task and achieve from 56.5 to 71.8 of F1 score using patient language, against 64.7 to 73.1 for the same task using medical language. When using patient language, we observe only a small loss in performance for the best model, suggesting that having the patient as a starting point could be adopted to help recruit patients for clinical trials. The corpus and code bases are all freely available on our Github and HuggingFace repositories.
SEME at SemEval-2024 Task 2: Comparing Masked and Generative Language Models on Natural Language Inference for Clinical Trials
Apr 05, 2024Abstract:This paper describes our submission to Task 2 of SemEval-2024: Safe Biomedical Natural Language Inference for Clinical Trials. The Multi-evidence Natural Language Inference for Clinical Trial Data (NLI4CT) consists of a Textual Entailment (TE) task focused on the evaluation of the consistency and faithfulness of Natural Language Inference (NLI) models applied to Clinical Trial Reports (CTR). We test 2 distinct approaches, one based on finetuning and ensembling Masked Language Models and the other based on prompting Large Language Models using templates, in particular, using Chain-Of-Thought and Contrastive Chain-Of-Thought. Prompting Flan-T5-large in a 2-shot setting leads to our best system that achieves 0.57 F1 score, 0.64 Faithfulness, and 0.56 Consistency.
A Dataset for Pharmacovigilance in German, French, and Japanese: Annotating Adverse Drug Reactions across Languages
Mar 27, 2024



Abstract:User-generated data sources have gained significance in uncovering Adverse Drug Reactions (ADRs), with an increasing number of discussions occurring in the digital world. However, the existing clinical corpora predominantly revolve around scientific articles in English. This work presents a multilingual corpus of texts concerning ADRs gathered from diverse sources, including patient fora, social media, and clinical reports in German, French, and Japanese. Our corpus contains annotations covering 12 entity types, four attribute types, and 13 relation types. It contributes to the development of real-world multilingual language models for healthcare. We provide statistics to highlight certain challenges associated with the corpus and conduct preliminary experiments resulting in strong baselines for extracting entities and relations between these entities, both within and across languages.
Cross-lingual Approaches for the Detection of Adverse Drug Reactions in German from a Patient's Perspective
Aug 03, 2022
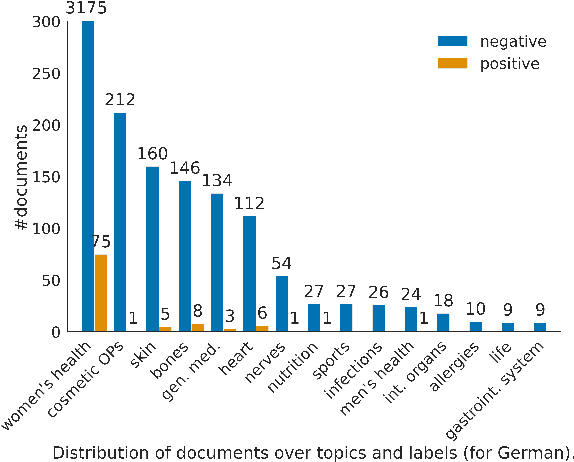
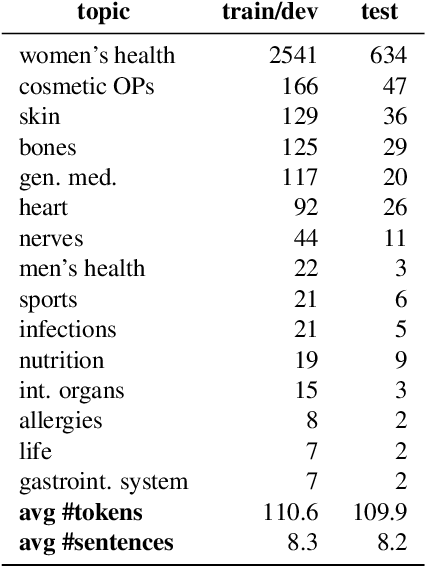
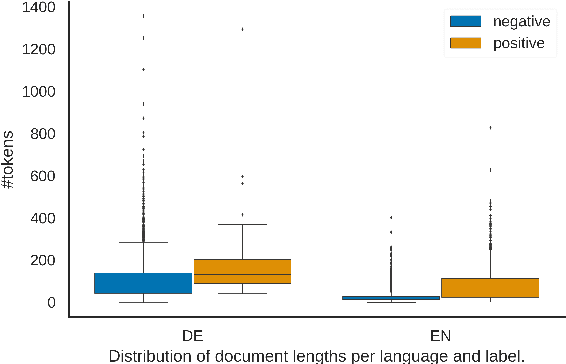
Abstract:In this work, we present the first corpus for German Adverse Drug Reaction (ADR) detection in patient-generated content. The data consists of 4,169 binary annotated documents from a German patient forum, where users talk about health issues and get advice from medical doctors. As is common in social media data in this domain, the class labels of the corpus are very imbalanced. This and a high topic imbalance make it a very challenging dataset, since often, the same symptom can have several causes and is not always related to a medication intake. We aim to encourage further multi-lingual efforts in the domain of ADR detection and provide preliminary experiments for binary classification using different methods of zero- and few-shot learning based on a multi-lingual model. When fine-tuning XLM-RoBERTa first on English patient forum data and then on the new German data, we achieve an F1-score of 37.52 for the positive class. We make the dataset and models publicly available for the community.
Decorate the Examples: A Simple Method of Prompt Design for Biomedical Relation Extraction
Apr 21, 2022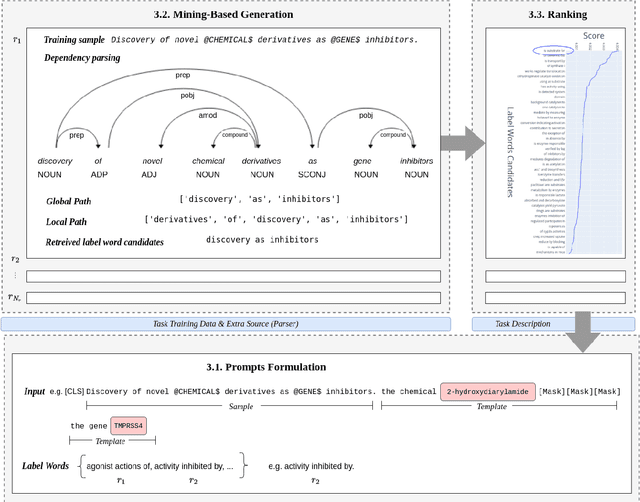
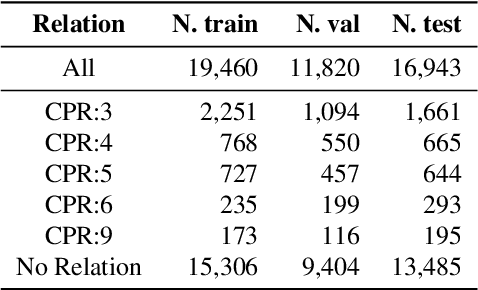
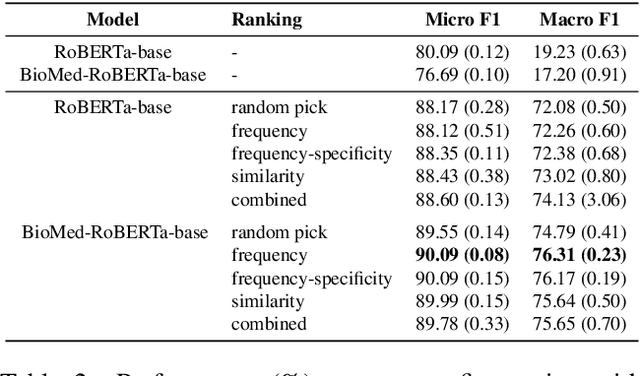
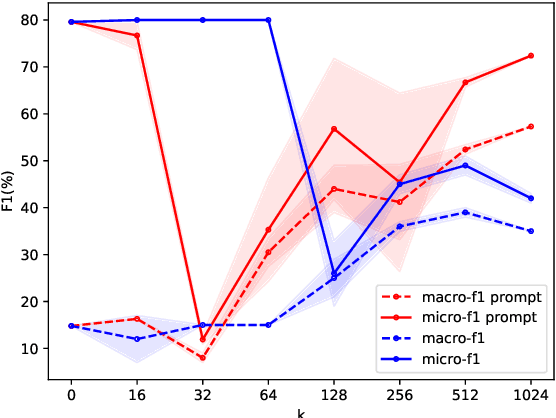
Abstract:Relation extraction is a core problem for natural language processing in the biomedical domain. Recent research on relation extraction showed that prompt-based learning improves the performance on both fine-tuning on full training set and few-shot training. However, less effort has been made on domain-specific tasks where good prompt design can be even harder. In this paper, we investigate prompting for biomedical relation extraction, with experiments on the ChemProt dataset. We present a simple yet effective method to systematically generate comprehensive prompts that reformulate the relation extraction task as a cloze-test task under a simple prompt formulation. In particular, we experiment with different ranking scores for prompt selection. With BioMed-RoBERTa-base, our results show that prompting-based fine-tuning obtains gains by 14.21 F1 over its regular fine-tuning baseline, and 1.14 F1 over SciFive-Large, the current state-of-the-art on ChemProt. Besides, we find prompt-based learning requires fewer training examples to make reasonable predictions. The results demonstrate the potential of our methods in such a domain-specific relation extraction task.
Does constituency analysis enhance domain-specific pre-trained BERT models for relation extraction?
Nov 25, 2021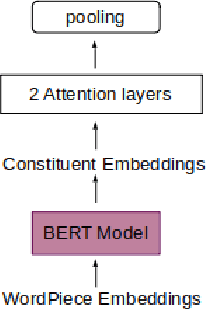
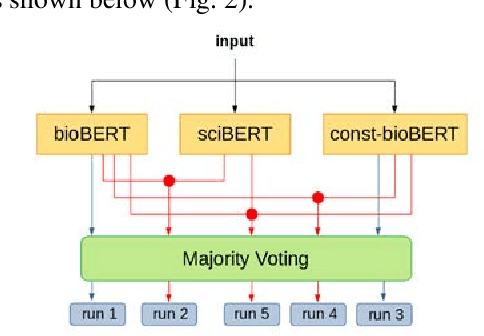
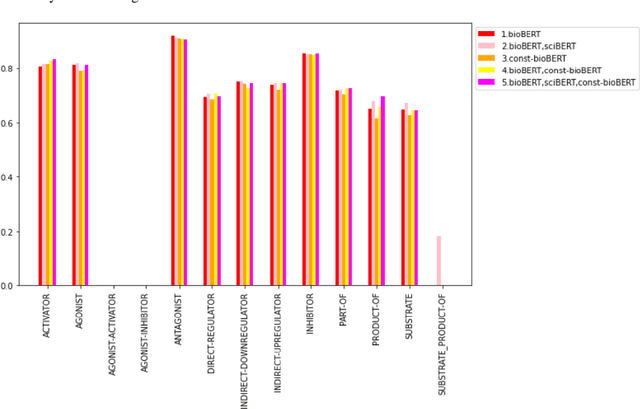
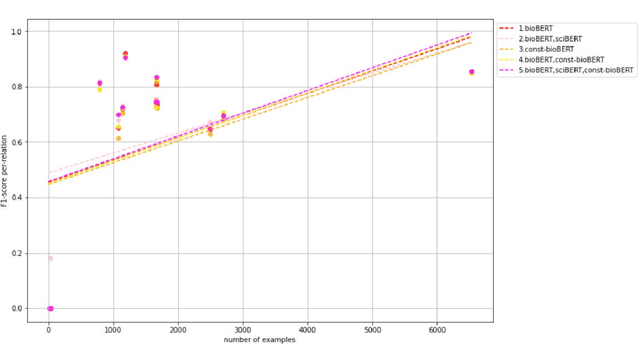
Abstract:Recently many studies have been conducted on the topic of relation extraction. The DrugProt track at BioCreative VII provides a manually-annotated corpus for the purpose of the development and evaluation of relation extraction systems, in which interactions between chemicals and genes are studied. We describe the ensemble system that we used for our submission, which combines predictions of fine-tuned bioBERT, sciBERT and const-bioBERT models by majority voting. We specifically tested the contribution of syntactic information to relation extraction with BERT. We observed that adding constituentbased syntactic information to BERT improved precision, but decreased recall, since relations rarely seen in the train set were less likely to be predicted by BERT models in which the syntactic information is infused. Our code is available online [https://github.com/Maple177/drugprot-relation-extraction].
Global alignment for relation extraction in Microbiology
Nov 25, 2021
Abstract:We investigate a method to extract relations from texts based on global alignment and syntactic information. Combined with SVM, this method is shown to have a performance comparable or even better than LSTM on two RE tasks.
CharacterBERT: Reconciling ELMo and BERT for Word-Level Open-Vocabulary Representations From Characters
Oct 31, 2020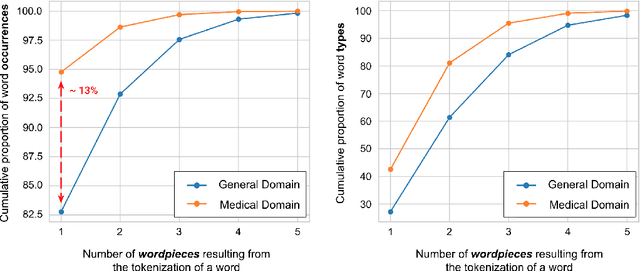

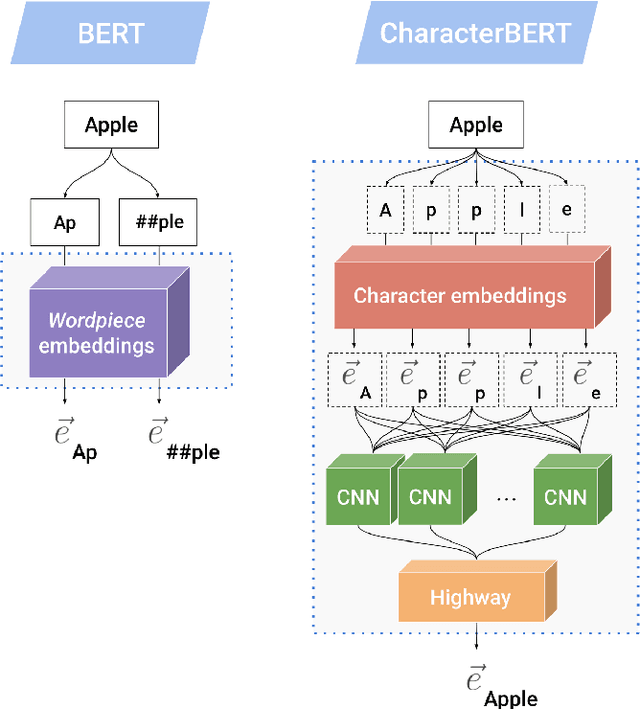
Abstract:Due to the compelling improvements brought by BERT, many recent representation models adopted the Transformer architecture as their main building block, consequently inheriting the wordpiece tokenization system despite it not being intrinsically linked to the notion of Transformers. While this system is thought to achieve a good balance between the flexibility of characters and the efficiency of full words, using predefined wordpiece vocabularies from the general domain is not always suitable, especially when building models for specialized domains (e.g., the medical domain). Moreover, adopting a wordpiece tokenization shifts the focus from the word level to the subword level, making the models conceptually more complex and arguably less convenient in practice. For these reasons, we propose CharacterBERT, a new variant of BERT that drops the wordpiece system altogether and uses a Character-CNN module instead to represent entire words by consulting their characters. We show that this new model improves the performance of BERT on a variety of medical domain tasks while at the same time producing robust, word-level and open-vocabulary representations.
Cross-Lingual Contextual Word Embeddings Mapping With Multi-Sense Words In Mind
Sep 18, 2019



Abstract:Recent work in cross-lingual contextual word embedding learning cannot handle multi-sense words well. In this work, we explore the characteristics of contextual word embeddings and show the link between contextual word embeddings and word senses. We propose two improving solutions by considering contextual multi-sense word embeddings as noise (removal) and by generating cluster level average anchor embeddings for contextual multi-sense word embeddings (replacement). Experiments show that our solutions can improve the supervised contextual word embeddings alignment for multi-sense words in a microscopic perspective without hurting the macroscopic performance on the bilingual lexicon induction task. For unsupervised alignment, our methods significantly improve the performance on the bilingual lexicon induction task for more than 10 points.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge