Nona Naderi
Collaborative Rational Speech Act: Pragmatic Reasoning for Multi-Turn Dialog
Jul 18, 2025Abstract:As AI systems take on collaborative roles, they must reason about shared goals and beliefs-not just generate fluent language. The Rational Speech Act (RSA) framework offers a principled approach to pragmatic reasoning, but existing extensions face challenges in scaling to multi-turn, collaborative scenarios. In this paper, we introduce Collaborative Rational Speech Act (CRSA), an information-theoretic (IT) extension of RSA that models multi-turn dialog by optimizing a gain function adapted from rate-distortion theory. This gain is an extension of the gain model that is maximized in the original RSA model but takes into account the scenario in which both agents in a conversation have private information and produce utterances conditioned on the dialog. We demonstrate the effectiveness of CRSA on referential games and template-based doctor-patient dialogs in the medical domain. Empirical results show that CRSA yields more consistent, interpretable, and collaborative behavior than existing baselines-paving the way for more pragmatic and socially aware language agents.
Am I eligible? Natural Language Inference for Clinical Trial Patient Recruitment: the Patient's Point of View
Mar 19, 2025



Abstract:Recruiting patients to participate in clinical trials can be challenging and time-consuming. Usually, participation in a clinical trial is initiated by a healthcare professional and proposed to the patient. Promoting clinical trials directly to patients via online recruitment might help to reach them more efficiently. In this study, we address the case where a patient is initiating their own recruitment process and wants to determine whether they are eligible for a given clinical trial, using their own language to describe their medical profile. To study whether this creates difficulties in the patient trial matching process, we design a new dataset and task, Natural Language Inference for Patient Recruitment (NLI4PR), in which patient language profiles must be matched to clinical trials. We create it by adapting the TREC 2022 Clinical Trial Track dataset, which provides patients' medical profiles, and rephrasing them manually using patient language. We also use the associated clinical trial reports where the patients are either eligible or excluded. We prompt several open-source Large Language Models on our task and achieve from 56.5 to 71.8 of F1 score using patient language, against 64.7 to 73.1 for the same task using medical language. When using patient language, we observe only a small loss in performance for the best model, suggesting that having the patient as a starting point could be adopted to help recruit patients for clinical trials. The corpus and code bases are all freely available on our Github and HuggingFace repositories.
SEME at SemEval-2024 Task 2: Comparing Masked and Generative Language Models on Natural Language Inference for Clinical Trials
Apr 05, 2024Abstract:This paper describes our submission to Task 2 of SemEval-2024: Safe Biomedical Natural Language Inference for Clinical Trials. The Multi-evidence Natural Language Inference for Clinical Trial Data (NLI4CT) consists of a Textual Entailment (TE) task focused on the evaluation of the consistency and faithfulness of Natural Language Inference (NLI) models applied to Clinical Trial Reports (CTR). We test 2 distinct approaches, one based on finetuning and ensembling Masked Language Models and the other based on prompting Large Language Models using templates, in particular, using Chain-Of-Thought and Contrastive Chain-Of-Thought. Prompting Flan-T5-large in a 2-shot setting leads to our best system that achieves 0.57 F1 score, 0.64 Faithfulness, and 0.56 Consistency.
DS4DH at TREC Health Misinformation 2021: Multi-Dimensional Ranking Models with Transfer Learning and Rank Fusion
Feb 14, 2022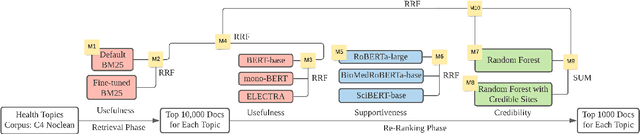
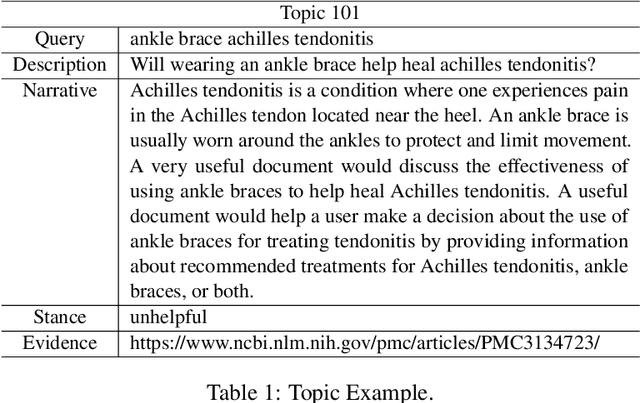
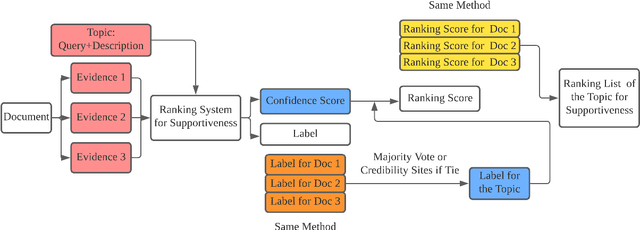
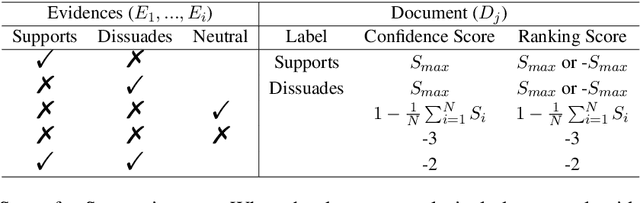
Abstract:This paper describes the work of the Data Science for Digital Health (DS4DH) group at the TREC Health Misinformation Track 2021. The TREC Health Misinformation track focused on the development of retrieval methods that provide relevant, correct and credible information for health related searches on the Web. In our methodology, we used a two-step ranking approach that includes i) a standard retrieval phase, based on BM25 model, and ii) a re-ranking phase, with a pipeline of models focused on the usefulness, supportiveness and credibility dimensions of the retrieved documents. To estimate the usefulness, we classified the initial rank list using pre-trained language models based on the transformers architecture fine-tuned on the MS MARCO corpus. To assess the supportiveness, we utilized BERT-based models fine-tuned on scientific and Wikipedia corpora. Finally, to evaluate the credibility of the documents, we employed a random forest model trained on the Microsoft Credibility dataset combined with a list of credible sites. The resulting ranked lists were then combined using the Reciprocal Rank Fusion algorithm to obtain the final list of useful, supporting and credible documents. Our approach achieved competitive results, being top-2 in the compatibility measurement for the automatic runs. Our findings suggest that integrating automatic ranking models created for each information quality dimension with transfer learning can increase the effectiveness of health-related information retrieval.
Named entity recognition in chemical patents using ensemble of contextual language models
Jul 24, 2020
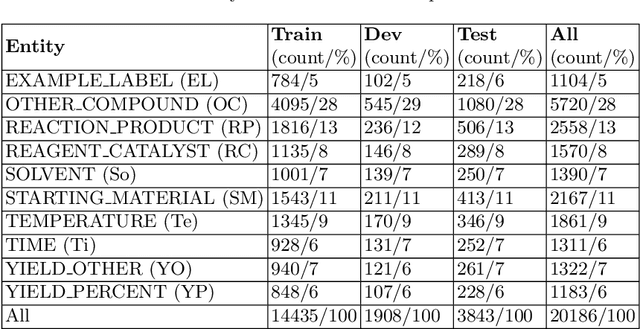
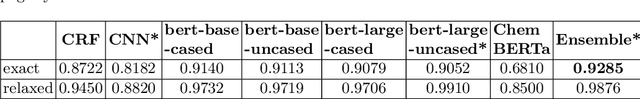
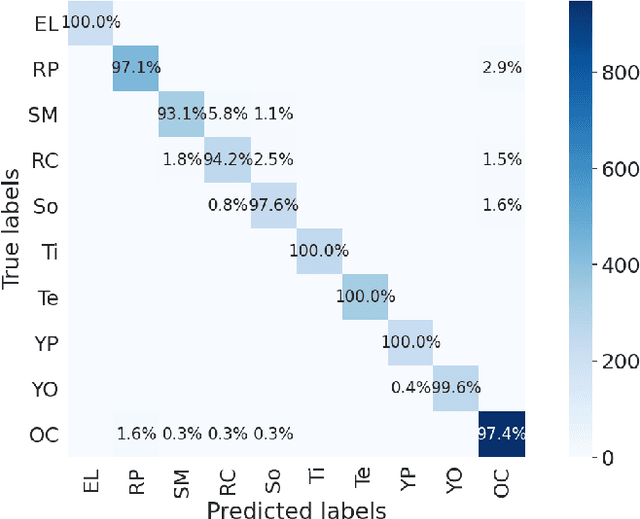
Abstract:Chemical patent documents describe a broad range of applications holding key information, such as chemical compounds, reactions, and specific properties. However, the key information should be enabled to be utilized in downstream tasks. Text mining provides means to extract relevant information from chemical patents through information extraction techniques. As part of the Information Extraction task of the Cheminformatics Elseiver Melbourne University challenge, in this work we study the effectiveness of contextualized language models to extract reaction information in chemical patents. We compare transformer architectures trained on a generic corpus with models specialised in chemistry patents, and propose a new model based on the combination of existing architectures. Our best model, based on the ensemble approach, achieves an exact F1-score of 92.30% and a relaxed F1 -score of 96.24%. We show that the ensemble of contextualized language models provides an effective method to extract information from chemical patents. As a next step, we will investigate the effect of transformer language models pre-trained in chemical patents.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge