Robert Bossy
LISN
Taec: a Manually annotated text dataset for trait and phenotype extraction and entity linking in wheat breeding literature
Jan 15, 2024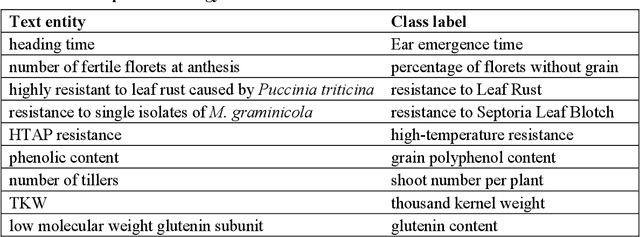
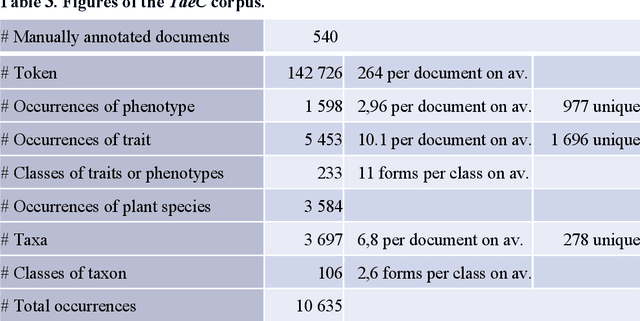
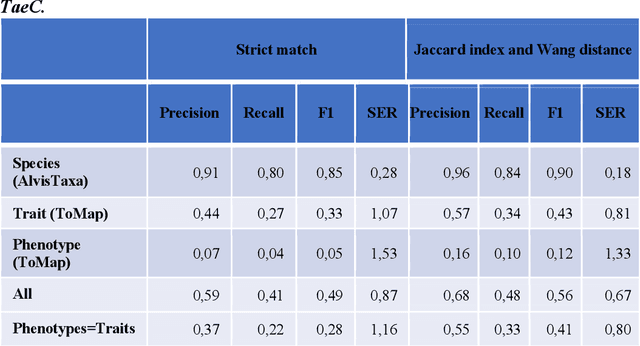
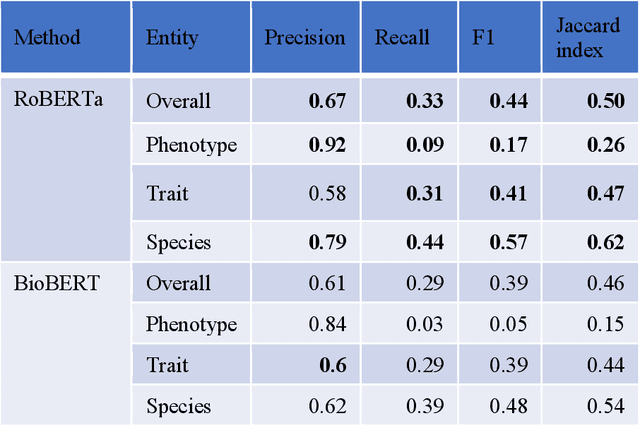
Abstract:Wheat varieties show a large diversity of traits and phenotypes. Linking them to genetic variability is essential for shorter and more efficient wheat breeding programs. Newly desirable wheat variety traits include disease resistance to reduce pesticide use, adaptation to climate change, resistance to heat and drought stresses, or low gluten content of grains. Wheat breeding experiments are documented by a large body of scientific literature and observational data obtained in-field and under controlled conditions. The cross-referencing of complementary information from the literature and observational data is essential to the study of the genotype-phenotype relationship and to the improvement of wheat selection. The scientific literature on genetic marker-assisted selection describes much information about the genotype-phenotype relationship. However, the variety of expressions used to refer to traits and phenotype values in scientific articles is a hinder to finding information and cross-referencing it. When trained adequately by annotated examples, recent text mining methods perform highly in named entity recognition and linking in the scientific domain. While several corpora contain annotations of human and animal phenotypes, currently, no corpus is available for training and evaluating named entity recognition and entity-linking methods in plant phenotype literature. The Triticum aestivum trait Corpus is a new gold standard for traits and phenotypes of wheat. It consists of 540 PubMed references fully annotated for trait, phenotype, and species named entities using the Wheat Trait and Phenotype Ontology and the species taxonomy of the National Center for Biotechnology Information. A study of the performance of tools trained on the Triticum aestivum trait Corpus shows that the corpus is suitable for the training and evaluation of named entity recognition and linking.
Does constituency analysis enhance domain-specific pre-trained BERT models for relation extraction?
Nov 25, 2021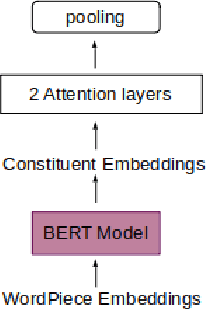
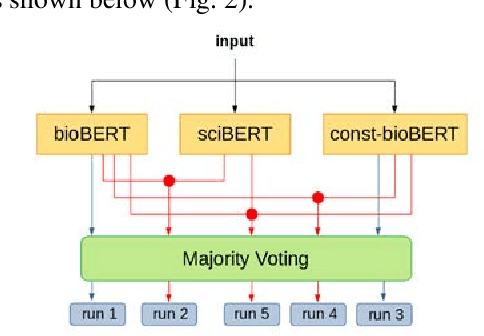
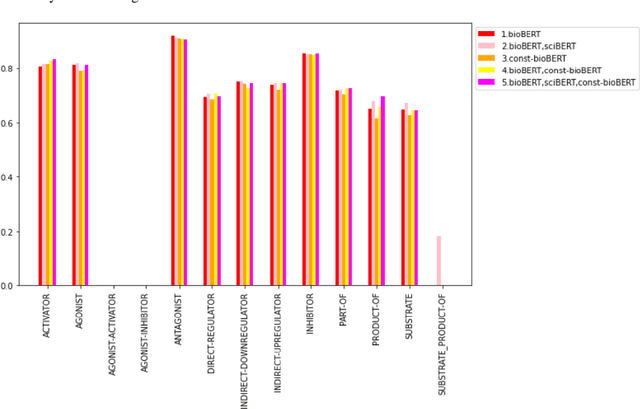
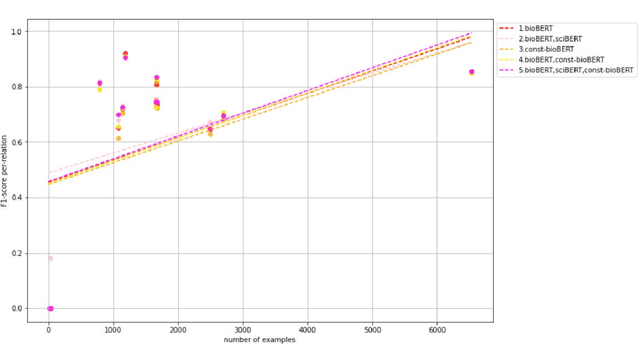
Abstract:Recently many studies have been conducted on the topic of relation extraction. The DrugProt track at BioCreative VII provides a manually-annotated corpus for the purpose of the development and evaluation of relation extraction systems, in which interactions between chemicals and genes are studied. We describe the ensemble system that we used for our submission, which combines predictions of fine-tuned bioBERT, sciBERT and const-bioBERT models by majority voting. We specifically tested the contribution of syntactic information to relation extraction with BERT. We observed that adding constituentbased syntactic information to BERT improved precision, but decreased recall, since relations rarely seen in the train set were less likely to be predicted by BERT models in which the syntactic information is infused. Our code is available online [https://github.com/Maple177/drugprot-relation-extraction].
Global alignment for relation extraction in Microbiology
Nov 25, 2021
Abstract:We investigate a method to extract relations from texts based on global alignment and syntactic information. Combined with SVM, this method is shown to have a performance comparable or even better than LSTM on two RE tasks.
Building Large Lexicalized Ontologies from Text: a Use Case in Automatic Indexing of Biotechnology Patents
Oct 02, 2020Abstract:This paper presents a tool, TyDI, and methods experimented in the building of a termino-ontology, i.e. a lexicalized ontology aimed at fine-grained indexation for semantic search applications. TyDI provides facilities for knowledge engineers and domain experts to efficiently collaborate to validate, organize and conceptualize corpus extracted terms. A use case on biotechnology patent search demonstrates TyDI's potential.
Text-mining and ontologies: new approaches to knowledge discovery of microbial diversity
Oct 31, 2018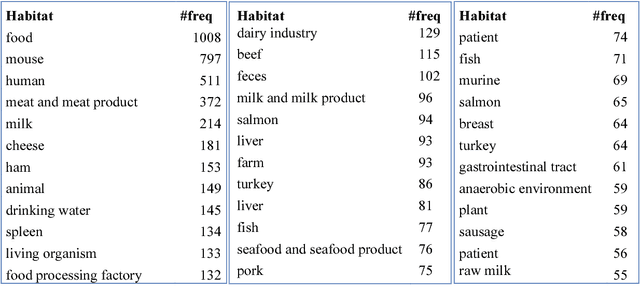
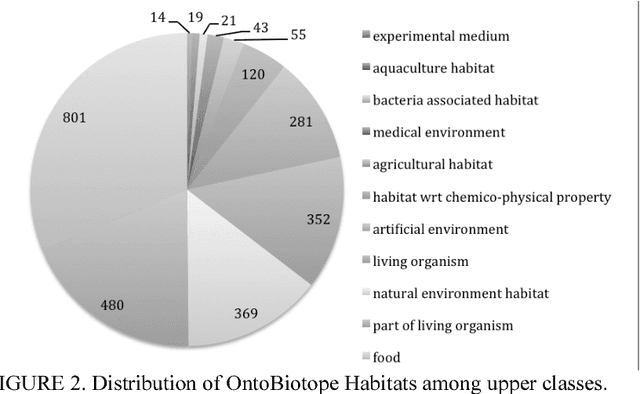
Abstract:Microbiology research has access to a very large amount of public information on the habitats of microorganisms. Many areas of microbiology research uses this information, primarily in biodiversity studies. However the habitat information is expressed in unstructured natural language form, which hinders its exploitation at large-scale. It is very common for similar habitats to be described by different terms, which makes them hard to compare automatically, e.g. intestine and gut. The use of a common reference to standardize these habitat descriptions as claimed by (Ivana et al., 2010) is a necessity. We propose the ontology called OntoBiotope that we have been developing since 2010. The OntoBiotope ontology is in a formal machine-readable representation that enables indexing of information as well as conceptualization and reasoning.
* 5 pages
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge