Mariya Borovikova
Algorithms For Automatic Accentuation And Transcription Of Russian Texts In Speech Recognition Systems
Oct 03, 2024Abstract:This paper presents an overview of rule-based system for automatic accentuation and phonemic transcription of Russian texts for speech connected tasks, such as Automatic Speech Recognition (ASR). Two parts of the developed system, accentuation and transcription, use different approaches to achieve correct phonemic representations of input phrases. Accentuation is based on "Grammatical dictionary of the Russian language" of A.A. Zaliznyak and wiktionary corpus. To distinguish homographs, the accentuation system also utilises morphological information of the sentences based on Recurrent Neural Networks (RNN). Transcription algorithms apply the rules presented in the monograph of B.M. Lobanov and L.I. Tsirulnik "Computer Synthesis and Voice Cloning". The rules described in the present paper are implemented in an open-source module, which can be of use to any scientific study connected to ASR or Speech To Text (STT) tasks. Automatically marked up text annotations of the Russian Voxforge database were used as training data for an acoustic model in CMU Sphinx. The resulting acoustic model was evaluated on cross-validation, mean Word Accuracy being 71.2%. The developed toolkit is written in the Python language and is accessible on GitHub for any researcher interested.
Taec: a Manually annotated text dataset for trait and phenotype extraction and entity linking in wheat breeding literature
Jan 15, 2024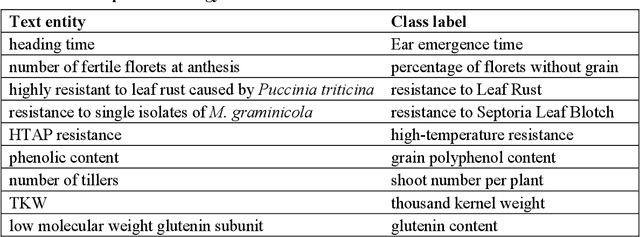
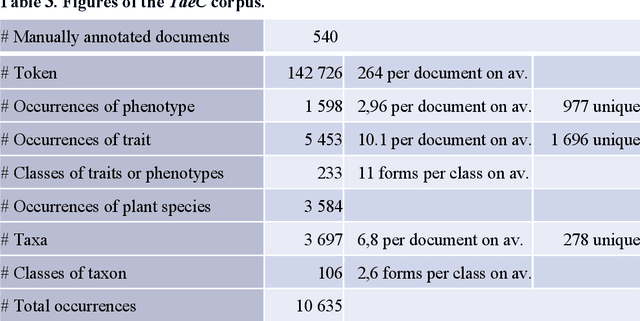
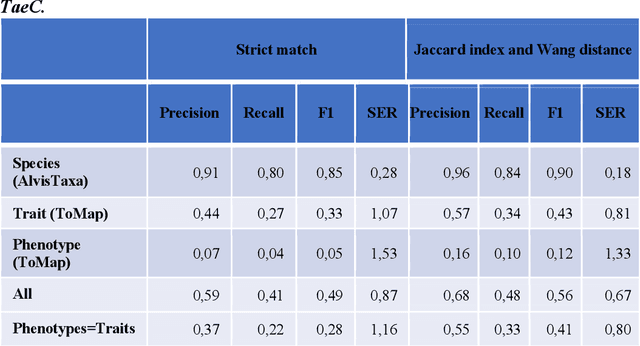
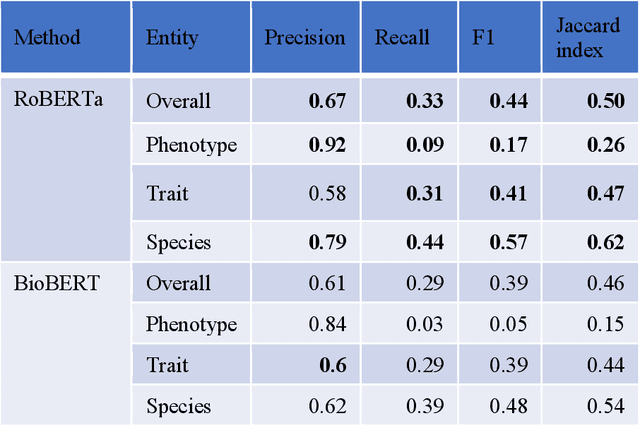
Abstract:Wheat varieties show a large diversity of traits and phenotypes. Linking them to genetic variability is essential for shorter and more efficient wheat breeding programs. Newly desirable wheat variety traits include disease resistance to reduce pesticide use, adaptation to climate change, resistance to heat and drought stresses, or low gluten content of grains. Wheat breeding experiments are documented by a large body of scientific literature and observational data obtained in-field and under controlled conditions. The cross-referencing of complementary information from the literature and observational data is essential to the study of the genotype-phenotype relationship and to the improvement of wheat selection. The scientific literature on genetic marker-assisted selection describes much information about the genotype-phenotype relationship. However, the variety of expressions used to refer to traits and phenotype values in scientific articles is a hinder to finding information and cross-referencing it. When trained adequately by annotated examples, recent text mining methods perform highly in named entity recognition and linking in the scientific domain. While several corpora contain annotations of human and animal phenotypes, currently, no corpus is available for training and evaluating named entity recognition and entity-linking methods in plant phenotype literature. The Triticum aestivum trait Corpus is a new gold standard for traits and phenotypes of wheat. It consists of 540 PubMed references fully annotated for trait, phenotype, and species named entities using the Wheat Trait and Phenotype Ontology and the species taxonomy of the National Center for Biotechnology Information. A study of the performance of tools trained on the Triticum aestivum trait Corpus shows that the corpus is suitable for the training and evaluation of named entity recognition and linking.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge