Danilo Schmidt
A Medical Information Extraction Workbench to Process German Clinical Text
Jul 08, 2022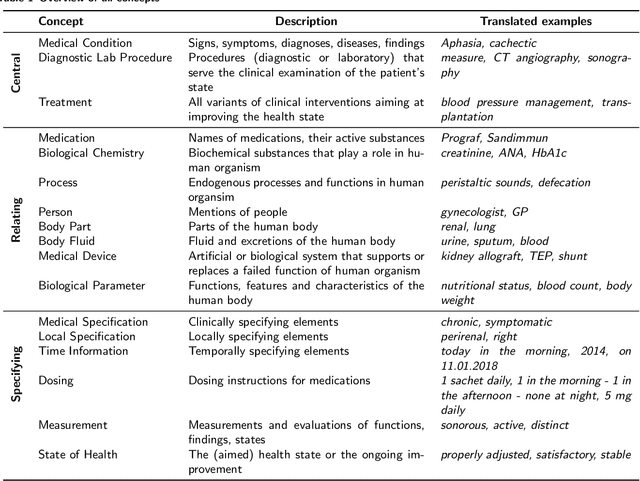
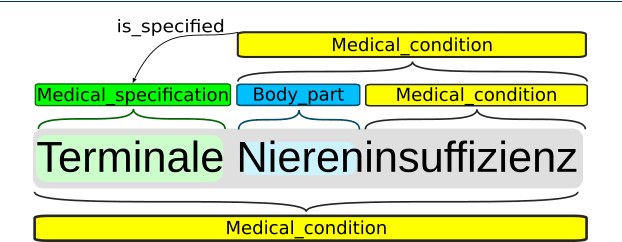

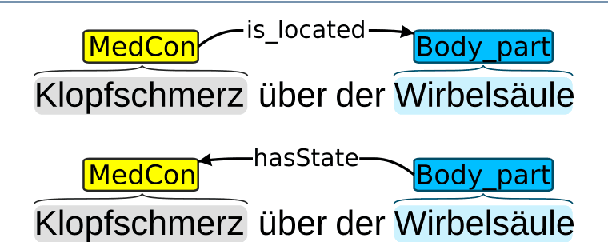
Abstract:Background: In the information extraction and natural language processing domain, accessible datasets are crucial to reproduce and compare results. Publicly available implementations and tools can serve as benchmark and facilitate the development of more complex applications. However, in the context of clinical text processing the number of accessible datasets is scarce -- and so is the number of existing tools. One of the main reasons is the sensitivity of the data. This problem is even more evident for non-English languages. Approach: In order to address this situation, we introduce a workbench: a collection of German clinical text processing models. The models are trained on a de-identified corpus of German nephrology reports. Result: The presented models provide promising results on in-domain data. Moreover, we show that our models can be also successfully applied to other biomedical text in German. Our workbench is made publicly available so it can be used out of the box, as a benchmark or transferred to related problems.
Towards a New Science of a Clinical Data Intelligence
Dec 30, 2013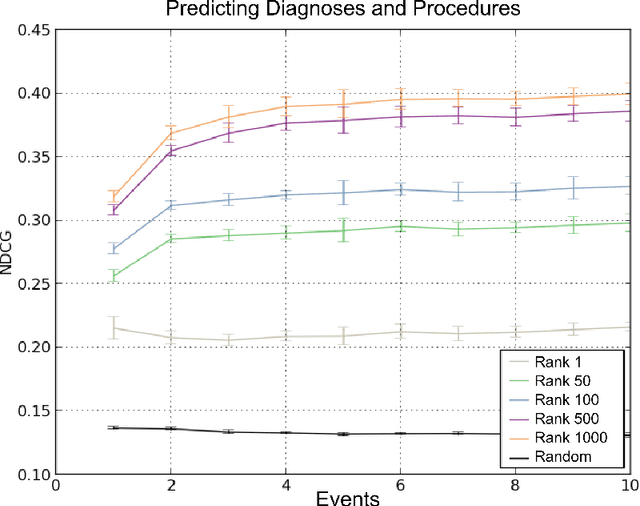
Abstract:In this paper we define Clinical Data Intelligence as the analysis of data generated in the clinical routine with the goal of improving patient care. We define a science of a Clinical Data Intelligence as a data analysis that permits the derivation of scientific, i.e., generalizable and reliable results. We argue that a science of a Clinical Data Intelligence is sensible in the context of a Big Data analysis, i.e., with data from many patients and with complete patient information. We discuss that Clinical Data Intelligence requires the joint efforts of knowledge engineering, information extraction (from textual and other unstructured data), and statistics and statistical machine learning. We describe some of our main results as conjectures and relate them to a recently funded research project involving two major German university hospitals.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge