Laura Seiffe
A Medical Information Extraction Workbench to Process German Clinical Text
Jul 08, 2022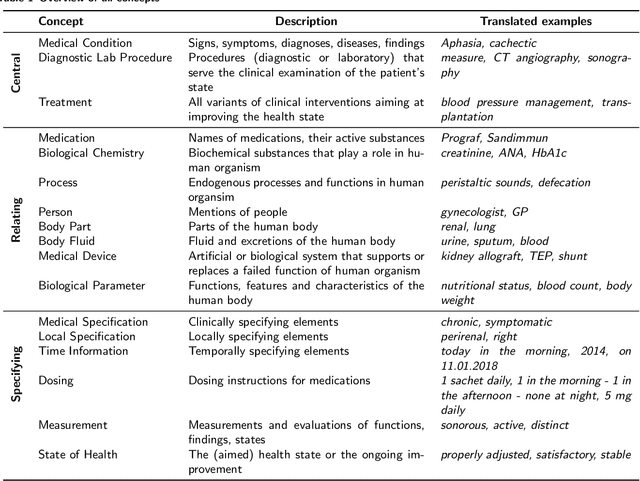
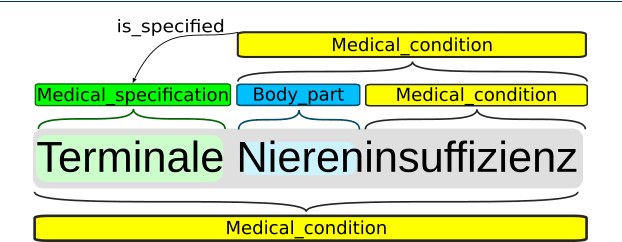

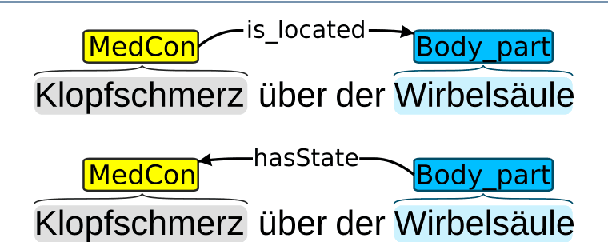
Abstract:Background: In the information extraction and natural language processing domain, accessible datasets are crucial to reproduce and compare results. Publicly available implementations and tools can serve as benchmark and facilitate the development of more complex applications. However, in the context of clinical text processing the number of accessible datasets is scarce -- and so is the number of existing tools. One of the main reasons is the sensitivity of the data. This problem is even more evident for non-English languages. Approach: In order to address this situation, we introduce a workbench: a collection of German clinical text processing models. The models are trained on a de-identified corpus of German nephrology reports. Result: The presented models provide promising results on in-domain data. Moreover, we show that our models can be also successfully applied to other biomedical text in German. Our workbench is made publicly available so it can be used out of the box, as a benchmark or transferred to related problems.
From Witch's Shot to Music Making Bones -- Resources for Medical Laymen to Technical Language and Vice Versa
May 23, 2020



Abstract:Many people share information in social media or forums, like food they eat, sports activities they do or events which have been visited. This also applies to information about a person's health status. Information we share online unveils directly or indirectly information about our lifestyle and health situation and thus provides a valuable data resource. If we can make advantage of that data, applications can be created that enable e.g. the detection of possible risk factors of diseases or adverse drug reactions of medications. However, as most people are not medical experts, language used might be more descriptive rather than the precise medical expression as medics do. To detect and use those relevant information, laymen language has to be translated and/or linked to the corresponding medical concept. This work presents baseline data sources in order to address this challenge for German. We introduce a new data set which annotates medical laymen and technical expressions in a patient forum, along with a set of medical synonyms and definitions, and present first baseline results on the data.
Making Neural QA as Simple as Possible but not Simpler
Jun 08, 2017



Abstract:Recent development of large-scale question answering (QA) datasets triggered a substantial amount of research into end-to-end neural architectures for QA. Increasingly complex systems have been conceived without comparison to simpler neural baseline systems that would justify their complexity. In this work, we propose a simple heuristic that guides the development of neural baseline systems for the extractive QA task. We find that there are two ingredients necessary for building a high-performing neural QA system: first, the awareness of question words while processing the context and second, a composition function that goes beyond simple bag-of-words modeling, such as recurrent neural networks. Our results show that FastQA, a system that meets these two requirements, can achieve very competitive performance compared with existing models. We argue that this surprising finding puts results of previous systems and the complexity of recent QA datasets into perspective.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge