Peter A. Fasching
Improved HER2 Tumor Segmentation with Subtype Balancing using Deep Generative Networks
Nov 11, 2022Abstract:Tumor segmentation in histopathology images is often complicated by its composition of different histological subtypes and class imbalance. Oversampling subtypes with low prevalence features is not a satisfactory solution since it eventually leads to overfitting. We propose to create synthetic images with semantically-conditioned deep generative networks and to combine subtype-balanced synthetic images with the original dataset to achieve better segmentation performance. We show the suitability of Generative Adversarial Networks (GANs) and especially diffusion models to create realistic images based on subtype-conditioning for the use case of HER2-stained histopathology. Additionally, we show the capability of diffusion models to conditionally inpaint HER2 tumor areas with modified subtypes. Combining the original dataset with the same amount of diffusion-generated images increased the tumor Dice score from 0.833 to 0.854 and almost halved the variance between the HER2 subtype recalls. These results create the basis for more reliable automatic HER2 analysis with lower performance variance between individual HER2 subtypes.
Categorical EHR Imputation with Generative Adversarial Nets
Aug 05, 2021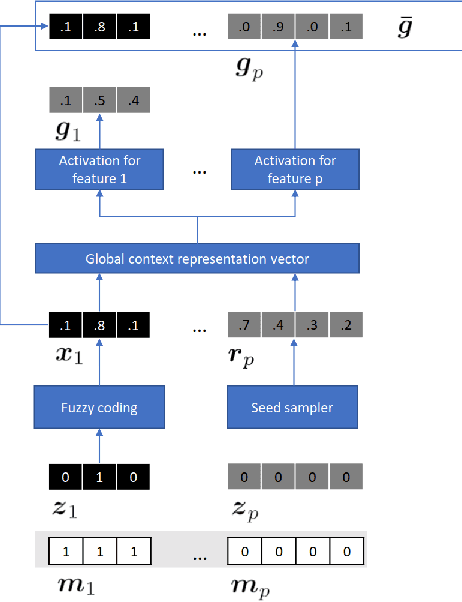
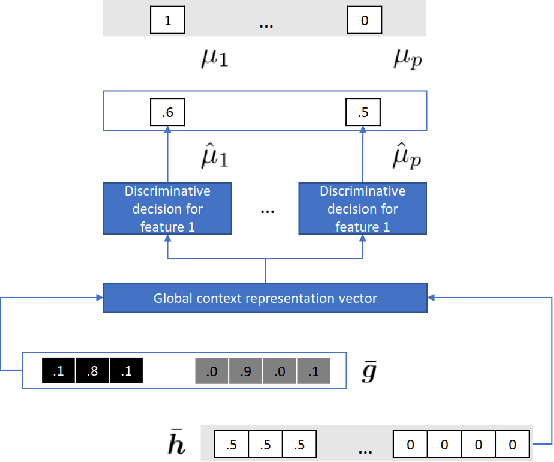
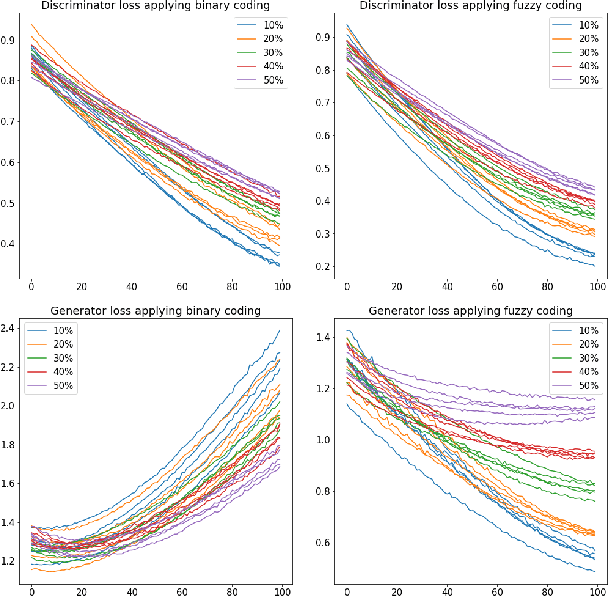
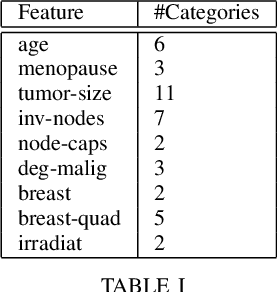
Abstract:Electronic Health Records often suffer from missing data, which poses a major problem in clinical practice and clinical studies. A novel approach for dealing with missing data are Generative Adversarial Nets (GANs), which have been generating huge research interest in image generation and transformation. Recently, researchers have attempted to apply GANs to missing data generation and imputation for EHR data: a major challenge here is the categorical nature of the data. State-of-the-art solutions to the GAN-based generation of categorical data involve either reinforcement learning, or learning a bidirectional mapping between the categorical and the real latent feature space, so that the GANs only need to generate real-valued features. However, these methods are designed to generate complete feature vectors instead of imputing only the subsets of missing features. In this paper we propose a simple and yet effective approach that is based on previous work on GANs for data imputation. We first motivate our solution by discussing the reason why adversarial training often fails in case of categorical features. Then we derive a novel way to re-code the categorical features to stabilize the adversarial training. Based on experiments on two real-world EHR data with multiple settings, we show that our imputation approach largely improves the prediction accuracy, compared to more traditional data imputation approaches.
Uncertainty-Aware Time-to-Event Prediction using Deep Kernel Accelerated Failure Time Models
Jul 26, 2021



Abstract:Recurrent neural network based solutions are increasingly being used in the analysis of longitudinal Electronic Health Record data. However, most works focus on prediction accuracy and neglect prediction uncertainty. We propose Deep Kernel Accelerated Failure Time models for the time-to-event prediction task, enabling uncertainty-awareness of the prediction by a pipeline of a recurrent neural network and a sparse Gaussian Process. Furthermore, a deep metric learning based pre-training step is adapted to enhance the proposed model. Our model shows better point estimate performance than recurrent neural network based baselines in experiments on two real-world datasets. More importantly, the predictive variance from our model can be used to quantify the uncertainty estimates of the time-to-event prediction: Our model delivers better performance when it is more confident in its prediction. Compared to related methods, such as Monte Carlo Dropout, our model offers better uncertainty estimates by leveraging an analytical solution and is more computationally efficient.
Predictive Clinical Decision Support System with RNN Encoding and Tensor Decoding
Dec 02, 2016


Abstract:With the introduction of the Electric Health Records, large amounts of digital data become available for analysis and decision support. When physicians are prescribing treatments to a patient, they need to consider a large range of data variety and volume, making decisions increasingly complex. Machine learning based Clinical Decision Support systems can be a solution to the data challenges. In this work we focus on a class of decision support in which the physicians' decision is directly predicted. Concretely, the model would assign higher probabilities to decisions that it presumes the physician are more likely to make. Thus the CDS system can provide physicians with rational recommendations. We also address the problem of correlation in target features: Often a physician is required to make multiple (sub-)decisions in a block, and that these decisions are mutually dependent. We propose a solution to the target correlation problem using a tensor factorization model. In order to handle the patients' historical information as sequential data, we apply the so-called Encoder-Decoder-Framework which is based on Recurrent Neural Networks (RNN) as encoders and a tensor factorization model as a decoder, a combination which is novel in machine learning. With experiments with real-world datasets we show that the proposed model does achieve better prediction performances.
Towards a New Science of a Clinical Data Intelligence
Dec 30, 2013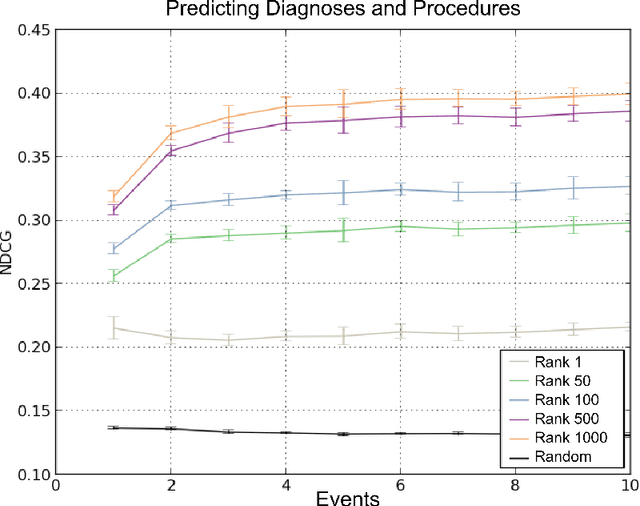
Abstract:In this paper we define Clinical Data Intelligence as the analysis of data generated in the clinical routine with the goal of improving patient care. We define a science of a Clinical Data Intelligence as a data analysis that permits the derivation of scientific, i.e., generalizable and reliable results. We argue that a science of a Clinical Data Intelligence is sensible in the context of a Big Data analysis, i.e., with data from many patients and with complete patient information. We discuss that Clinical Data Intelligence requires the joint efforts of knowledge engineering, information extraction (from textual and other unstructured data), and statistics and statistical machine learning. We describe some of our main results as conjectures and relate them to a recently funded research project involving two major German university hospitals.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge