Matthias Rübner
Style-Extracting Diffusion Models for Semi-Supervised Histopathology Segmentation
Mar 21, 2024Abstract:Deep learning-based image generation has seen significant advancements with diffusion models, notably improving the quality of generated images. Despite these developments, generating images with unseen characteristics beneficial for downstream tasks has received limited attention. To bridge this gap, we propose Style-Extracting Diffusion Models, featuring two conditioning mechanisms. Specifically, we utilize 1) a style conditioning mechanism which allows to inject style information of previously unseen images during image generation and 2) a content conditioning which can be targeted to a downstream task, e.g., layout for segmentation. We introduce a trainable style encoder to extract style information from images, and an aggregation block that merges style information from multiple style inputs. This architecture enables the generation of images with unseen styles in a zero-shot manner, by leveraging styles from unseen images, resulting in more diverse generations. In this work, we use the image layout as target condition and first show the capability of our method on a natural image dataset as a proof-of-concept. We further demonstrate its versatility in histopathology, where we combine prior knowledge about tissue composition and unannotated data to create diverse synthetic images with known layouts. This allows us to generate additional synthetic data to train a segmentation network in a semi-supervised fashion. We verify the added value of the generated images by showing improved segmentation results and lower performance variability between patients when synthetic images are included during segmentation training. Our code will be made publicly available at [LINK].
Analysing Diffusion Segmentation for Medical Images
Mar 21, 2024



Abstract:Denoising Diffusion Probabilistic models have become increasingly popular due to their ability to offer probabilistic modeling and generate diverse outputs. This versatility inspired their adaptation for image segmentation, where multiple predictions of the model can produce segmentation results that not only achieve high quality but also capture the uncertainty inherent in the model. Here, powerful architectures were proposed for improving diffusion segmentation performance. However, there is a notable lack of analysis and discussions on the differences between diffusion segmentation and image generation, and thorough evaluations are missing that distinguish the improvements these architectures provide for segmentation in general from their benefit for diffusion segmentation specifically. In this work, we critically analyse and discuss how diffusion segmentation for medical images differs from diffusion image generation, with a particular focus on the training behavior. Furthermore, we conduct an assessment how proposed diffusion segmentation architectures perform when trained directly for segmentation. Lastly, we explore how different medical segmentation tasks influence the diffusion segmentation behavior and the diffusion process could be adapted accordingly. With these analyses, we aim to provide in-depth insights into the behavior of diffusion segmentation that allow for a better design and evaluation of diffusion segmentation methods in the future.
Improved HER2 Tumor Segmentation with Subtype Balancing using Deep Generative Networks
Nov 11, 2022Abstract:Tumor segmentation in histopathology images is often complicated by its composition of different histological subtypes and class imbalance. Oversampling subtypes with low prevalence features is not a satisfactory solution since it eventually leads to overfitting. We propose to create synthetic images with semantically-conditioned deep generative networks and to combine subtype-balanced synthetic images with the original dataset to achieve better segmentation performance. We show the suitability of Generative Adversarial Networks (GANs) and especially diffusion models to create realistic images based on subtype-conditioning for the use case of HER2-stained histopathology. Additionally, we show the capability of diffusion models to conditionally inpaint HER2 tumor areas with modified subtypes. Combining the original dataset with the same amount of diffusion-generated images increased the tumor Dice score from 0.833 to 0.854 and almost halved the variance between the HER2 subtype recalls. These results create the basis for more reliable automatic HER2 analysis with lower performance variance between individual HER2 subtypes.
Superpixel Pre-Segmentation of HER2 Slides for Efficient Annotation
Jan 19, 2022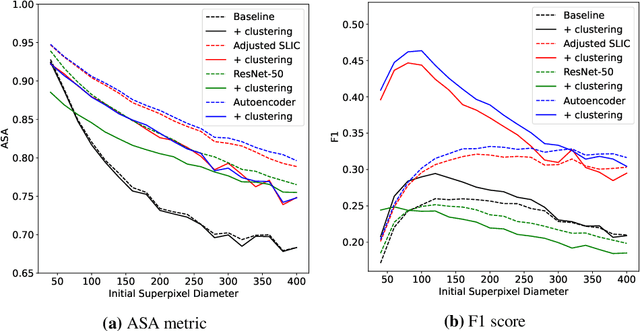
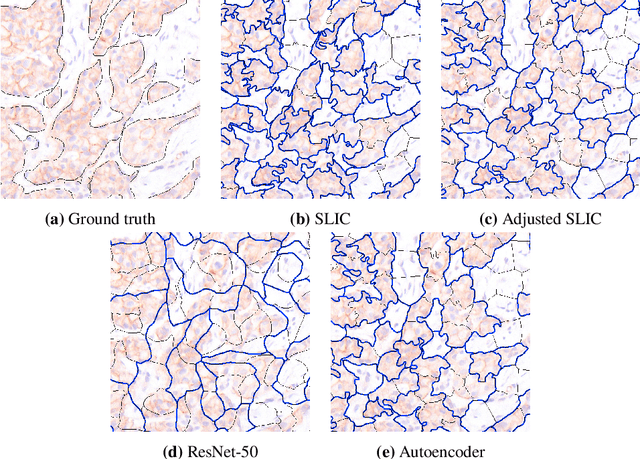
Abstract:Supervised deep learning has shown state-of-the-art performance for medical image segmentation across different applications, including histopathology and cancer research; however, the manual annotation of such data is extremely laborious. In this work, we explore the use of superpixel approaches to compute a pre-segmentation of HER2 stained images for breast cancer diagnosis that facilitates faster manual annotation and correction in a second step. Four methods are compared: Standard Simple Linear Iterative Clustering (SLIC) as a baseline, a domain adapted SLIC, and superpixels based on feature embeddings of a pretrained ResNet-50 and a denoising autoencoder. To tackle oversegmentation, we propose to hierarchically merge superpixels, based on their content in the respective feature space. When evaluating the approaches on fully manually annotated images, we observe that the autoencoder-based superpixels achieve a 23% increase in boundary F1 score compared to the baseline SLIC superpixels. Furthermore, the boundary F1 score increases by 73% when hierarchical clustering is applied on the adapted SLIC and the autoencoder-based superpixels. These evaluations show encouraging first results for a pre-segmentation for efficient manual refinement without the need for an initial set of annotated training data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge