Yinchong Yang
Fine-Grained Uncertainty Decomposition in Large Language Models: A Spectral Approach
Sep 26, 2025Abstract:As Large Language Models (LLMs) are increasingly integrated in diverse applications, obtaining reliable measures of their predictive uncertainty has become critically important. A precise distinction between aleatoric uncertainty, arising from inherent ambiguities within input data, and epistemic uncertainty, originating exclusively from model limitations, is essential to effectively address each uncertainty source. In this paper, we introduce Spectral Uncertainty, a novel approach to quantifying and decomposing uncertainties in LLMs. Leveraging the Von Neumann entropy from quantum information theory, Spectral Uncertainty provides a rigorous theoretical foundation for separating total uncertainty into distinct aleatoric and epistemic components. Unlike existing baseline methods, our approach incorporates a fine-grained representation of semantic similarity, enabling nuanced differentiation among various semantic interpretations in model responses. Empirical evaluations demonstrate that Spectral Uncertainty outperforms state-of-the-art methods in estimating both aleatoric and total uncertainty across diverse models and benchmark datasets.
Categorical EHR Imputation with Generative Adversarial Nets
Aug 05, 2021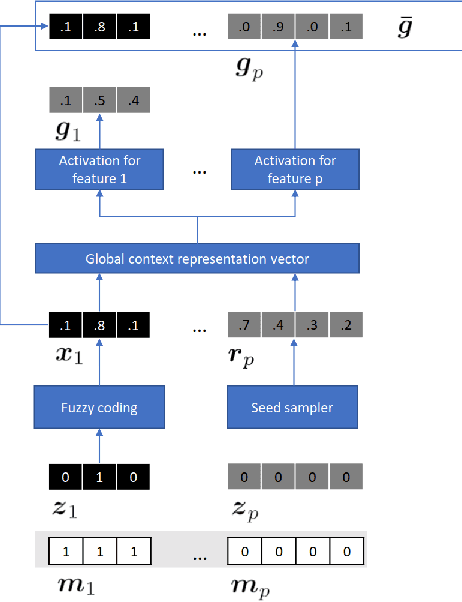
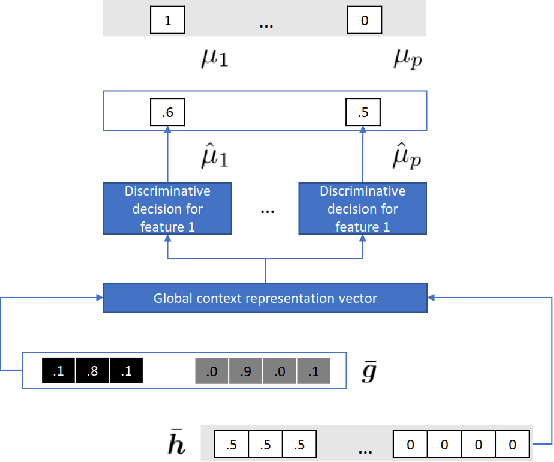
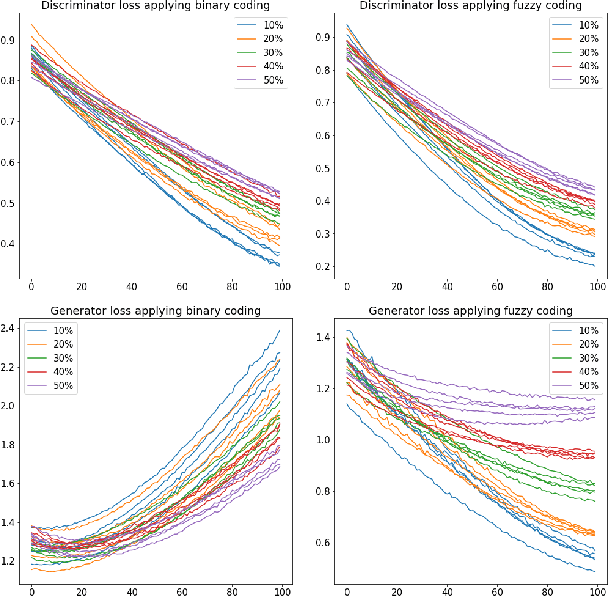
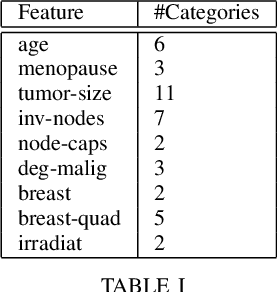
Abstract:Electronic Health Records often suffer from missing data, which poses a major problem in clinical practice and clinical studies. A novel approach for dealing with missing data are Generative Adversarial Nets (GANs), which have been generating huge research interest in image generation and transformation. Recently, researchers have attempted to apply GANs to missing data generation and imputation for EHR data: a major challenge here is the categorical nature of the data. State-of-the-art solutions to the GAN-based generation of categorical data involve either reinforcement learning, or learning a bidirectional mapping between the categorical and the real latent feature space, so that the GANs only need to generate real-valued features. However, these methods are designed to generate complete feature vectors instead of imputing only the subsets of missing features. In this paper we propose a simple and yet effective approach that is based on previous work on GANs for data imputation. We first motivate our solution by discussing the reason why adversarial training often fails in case of categorical features. Then we derive a novel way to re-code the categorical features to stabilize the adversarial training. Based on experiments on two real-world EHR data with multiple settings, we show that our imputation approach largely improves the prediction accuracy, compared to more traditional data imputation approaches.
Uncertainty-Aware Time-to-Event Prediction using Deep Kernel Accelerated Failure Time Models
Jul 26, 2021



Abstract:Recurrent neural network based solutions are increasingly being used in the analysis of longitudinal Electronic Health Record data. However, most works focus on prediction accuracy and neglect prediction uncertainty. We propose Deep Kernel Accelerated Failure Time models for the time-to-event prediction task, enabling uncertainty-awareness of the prediction by a pipeline of a recurrent neural network and a sparse Gaussian Process. Furthermore, a deep metric learning based pre-training step is adapted to enhance the proposed model. Our model shows better point estimate performance than recurrent neural network based baselines in experiments on two real-world datasets. More importantly, the predictive variance from our model can be used to quantify the uncertainty estimates of the time-to-event prediction: Our model delivers better performance when it is more confident in its prediction. Compared to related methods, such as Monte Carlo Dropout, our model offers better uncertainty estimates by leveraging an analytical solution and is more computationally efficient.
Multi-output Gaussian Processes for Uncertainty-aware Recommender Systems
Jun 08, 2021



Abstract:Recommender systems are often designed based on a collaborative filtering approach, where user preferences are predicted by modelling interactions between users and items. Many common approaches to solve the collaborative filtering task are based on learning representations of users and items, including simple matrix factorization, Gaussian process latent variable models, and neural-network based embeddings. While matrix factorization approaches fail to model nonlinear relations, neural networks can potentially capture such complex relations with unprecedented predictive power and are highly scalable. However, neither of them is able to model predictive uncertainties. In contrast, Gaussian Process based models can generate a predictive distribution, but cannot scale to large amounts of data. In this manuscript, we propose a novel approach combining the representation learning paradigm of collaborative filtering with multi-output Gaussian processes in a joint framework to generate uncertainty-aware recommendations. We introduce an efficient strategy for model training and inference, resulting in a model that scales to very large and sparse datasets and achieves competitive performance in terms of classical metrics quantifying the reconstruction error. In addition to accurately predicting user preferences, our model also provides meaningful uncertainty estimates about that prediction.
Quantifying Predictive Uncertainty in Medical Image Analysis with Deep Kernel Learning
Jun 01, 2021



Abstract:Deep neural networks are increasingly being used for the analysis of medical images. However, most works neglect the uncertainty in the model's prediction. We propose an uncertainty-aware deep kernel learning model which permits the estimation of the uncertainty in the prediction by a pipeline of a Convolutional Neural Network and a sparse Gaussian Process. Furthermore, we adapt different pre-training methods to investigate their impacts on the proposed model. We apply our approach to Bone Age Prediction and Lesion Localization. In most cases, the proposed model shows better performance compared to common architectures. More importantly, our model expresses systematically higher confidence in more accurate predictions and less confidence in less accurate ones. Our model can also be used to detect challenging and controversial test samples. Compared to related methods such as Monte-Carlo Dropout, our approach derives the uncertainty information in a purely analytical fashion and is thus computationally more efficient.
Learning Individualized Treatment Rules with Estimated Translated Inverse Propensity Score
Jul 02, 2020



Abstract:Randomized controlled trials typically analyze the effectiveness of treatments with the goal of making treatment recommendations for patient subgroups. With the advance of electronic health records, a great variety of data has been collected in clinical practice, enabling the evaluation of treatments and treatment policies based on observational data. In this paper, we focus on learning individualized treatment rules (ITRs) to derive a treatment policy that is expected to generate a better outcome for an individual patient. In our framework, we cast ITRs learning as a contextual bandit problem and minimize the expected risk of the treatment policy. We conduct experiments with the proposed framework both in a simulation study and based on a real-world dataset. In the latter case, we apply our proposed method to learn the optimal ITRs for the administration of intravenous (IV) fluids and vasopressors (VP). Based on various offline evaluation methods, we could show that the policy derived in our framework demonstrates better performance compared to both the physicians and other baselines, including a simple treatment prediction approach. As a long-term goal, our derived policy might eventually lead to better clinical guidelines for the administration of IV and VP.
Understanding Individual Decisions of CNNs via Contrastive Backpropagation
Dec 05, 2018



Abstract:A number of backpropagation-based approaches such as DeConvNets, vanilla Gradient Visualization and Guided Backpropagation have been proposed to better understand individual decisions of deep convolutional neural networks. The saliency maps produced by them are proven to be non-discriminative. Recently, the Layer-wise Relevance Propagation (LRP) approach was proposed to explain the classification decisions of rectifier neural networks. In this work, we evaluate the discriminativeness of the generated explanations and analyze the theoretical foundation of LRP, i.e. Deep Taylor Decomposition. The experiments and analysis conclude that the explanations generated by LRP are not class-discriminative. Based on LRP, we propose Contrastive Layer-wise Relevance Propagation (CLRP), which is capable of producing instance-specific, class-discriminative, pixel-wise explanations. In the experiments, we use the CLRP to explain the decisions and understand the difference between neurons in individual classification decisions. We also evaluate the explanations quantitatively with a Pointing Game and an ablation study. Both qualitative and quantitative evaluations show that the CLRP generates better explanations than the LRP.
* 16 pages, 5 figures, ACCV
Tensor-Train Recurrent Neural Networks for Video Classification
Jul 06, 2017
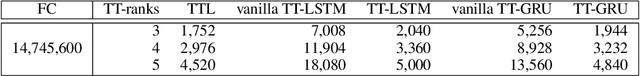
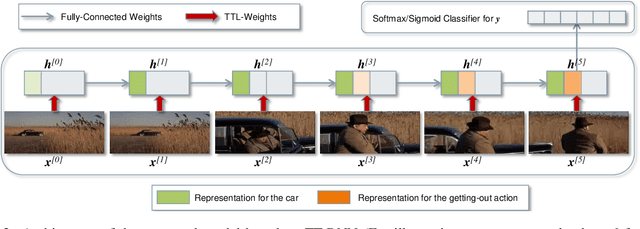
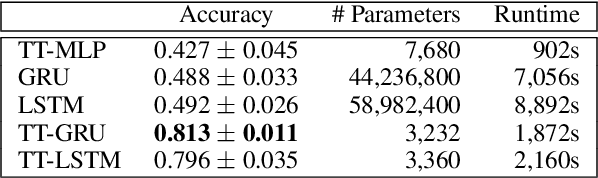
Abstract:The Recurrent Neural Networks and their variants have shown promising performances in sequence modeling tasks such as Natural Language Processing. These models, however, turn out to be impractical and difficult to train when exposed to very high-dimensional inputs due to the large input-to-hidden weight matrix. This may have prevented RNNs' large-scale application in tasks that involve very high input dimensions such as video modeling; current approaches reduce the input dimensions using various feature extractors. To address this challenge, we propose a new, more general and efficient approach by factorizing the input-to-hidden weight matrix using Tensor-Train decomposition which is trained simultaneously with the weights themselves. We test our model on classification tasks using multiple real-world video datasets and achieve competitive performances with state-of-the-art models, even though our model architecture is orders of magnitude less complex. We believe that the proposed approach provides a novel and fundamental building block for modeling high-dimensional sequential data with RNN architectures and opens up many possibilities to transfer the expressive and advanced architectures from other domains such as NLP to modeling high-dimensional sequential data.
Predictive Clinical Decision Support System with RNN Encoding and Tensor Decoding
Dec 02, 2016


Abstract:With the introduction of the Electric Health Records, large amounts of digital data become available for analysis and decision support. When physicians are prescribing treatments to a patient, they need to consider a large range of data variety and volume, making decisions increasingly complex. Machine learning based Clinical Decision Support systems can be a solution to the data challenges. In this work we focus on a class of decision support in which the physicians' decision is directly predicted. Concretely, the model would assign higher probabilities to decisions that it presumes the physician are more likely to make. Thus the CDS system can provide physicians with rational recommendations. We also address the problem of correlation in target features: Often a physician is required to make multiple (sub-)decisions in a block, and that these decisions are mutually dependent. We propose a solution to the target correlation problem using a tensor factorization model. In order to handle the patients' historical information as sequential data, we apply the so-called Encoder-Decoder-Framework which is based on Recurrent Neural Networks (RNN) as encoders and a tensor factorization model as a decoder, a combination which is novel in machine learning. With experiments with real-world datasets we show that the proposed model does achieve better prediction performances.
Predicting Clinical Events by Combining Static and Dynamic Information Using Recurrent Neural Networks
Nov 17, 2016



Abstract:In clinical data sets we often find static information (e.g. patient gender, blood type, etc.) combined with sequences of data that are recorded during multiple hospital visits (e.g. medications prescribed, tests performed, etc.). Recurrent Neural Networks (RNNs) have proven to be very successful for modelling sequences of data in many areas of Machine Learning. In this work we present an approach based on RNNs, specifically designed for the clinical domain, that combines static and dynamic information in order to predict future events. We work with a database collected in the Charit\'{e} Hospital in Berlin that contains complete information concerning patients that underwent a kidney transplantation. After the transplantation three main endpoints can occur: rejection of the kidney, loss of the kidney and death of the patient. Our goal is to predict, based on information recorded in the Electronic Health Record of each patient, whether any of those endpoints will occur within the next six or twelve months after each visit to the clinic. We compared different types of RNNs that we developed for this work, with a model based on a Feedforward Neural Network and a Logistic Regression model. We found that the RNN that we developed based on Gated Recurrent Units provides the best performance for this task. We also used the same models for a second task, i.e., next event prediction, and found that here the model based on a Feedforward Neural Network outperformed the other models. Our hypothesis is that long-term dependencies are not as relevant in this task.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge