Alexander Meyer
Infherno: End-to-end Agent-based FHIR Resource Synthesis from Free-form Clinical Notes
Jul 16, 2025Abstract:For clinical data integration and healthcare services, the HL7 FHIR standard has established itself as a desirable format for interoperability between complex health data. Previous attempts at automating the translation from free-form clinical notes into structured FHIR resources rely on modular, rule-based systems or LLMs with instruction tuning and constrained decoding. Since they frequently suffer from limited generalizability and structural inconformity, we propose an end-to-end framework powered by LLM agents, code execution, and healthcare terminology database tools to address these issues. Our solution, called Infherno, is designed to adhere to the FHIR document schema and competes well with a human baseline in predicting FHIR resources from unstructured text. The implementation features a front end for custom and synthetic data and both local and proprietary models, supporting clinical data integration processes and interoperability across institutions.
Beyond One-Time Validation: A Framework for Adaptive Validation of Prognostic and Diagnostic AI-based Medical Devices
Sep 07, 2024Abstract:Prognostic and diagnostic AI-based medical devices hold immense promise for advancing healthcare, yet their rapid development has outpaced the establishment of appropriate validation methods. Existing approaches often fall short in addressing the complexity of practically deploying these devices and ensuring their effective, continued operation in real-world settings. Building on recent discussions around the validation of AI models in medicine and drawing from validation practices in other fields, a framework to address this gap is presented. It offers a structured, robust approach to validation that helps ensure device reliability across differing clinical environments. The primary challenges to device performance upon deployment are discussed while highlighting the impact of changes related to individual healthcare institutions and operational processes. The presented framework emphasizes the importance of repeating validation and fine-tuning during deployment, aiming to mitigate these issues while being adaptable to challenges unforeseen during device development. The framework is also positioned within the current US and EU regulatory landscapes, underscoring its practical viability and relevance considering regulatory requirements. Additionally, a practical example demonstrating potential benefits of the framework is presented. Lastly, guidance on assessing model performance is offered and the importance of involving clinical stakeholders in the validation and fine-tuning process is discussed.
Multi-Modal Dataset Creation for Federated~Learning with DICOM Structured Reports
Jul 12, 2024



Abstract:Purpose: Federated training is often hindered by heterogeneous datasets due to divergent data storage options, inconsistent naming schemes, varied annotation procedures, and disparities in label quality. This is particularly evident in the emerging multi-modal learning paradigms, where dataset harmonization including a uniform data representation and filtering options are of paramount importance. Methods: DICOM structured reports enable the standardized linkage of arbitrary information beyond the imaging domain and can be used within Python deep learning pipelines with highdicom. Building on this, we developed an open platform for data integration and interactive filtering capabilities that simplifies the process of assembling multi-modal datasets. Results: In this study, we extend our prior work by showing its applicability to more and divergent data types, as well as streamlining datasets for federated training within an established consortium of eight university hospitals in Germany. We prove its concurrent filtering ability by creating harmonized multi-modal datasets across all locations for predicting the outcome after minimally invasive heart valve replacement. The data includes DICOM data (i.e. computed tomography images, electrocardiography scans) as well as annotations (i.e. calcification segmentations, pointsets and pacemaker dependency), and metadata (i.e. prosthesis and diagnoses). Conclusion: Structured reports bridge the traditional gap between imaging systems and information systems. Utilizing the inherent DICOM reference system arbitrary data types can be queried concurrently to create meaningful cohorts for clinical studies. The graphical interface as well as example structured report templates will be made publicly available.
Federated Foundation Model for Cardiac CT Imaging
Jul 10, 2024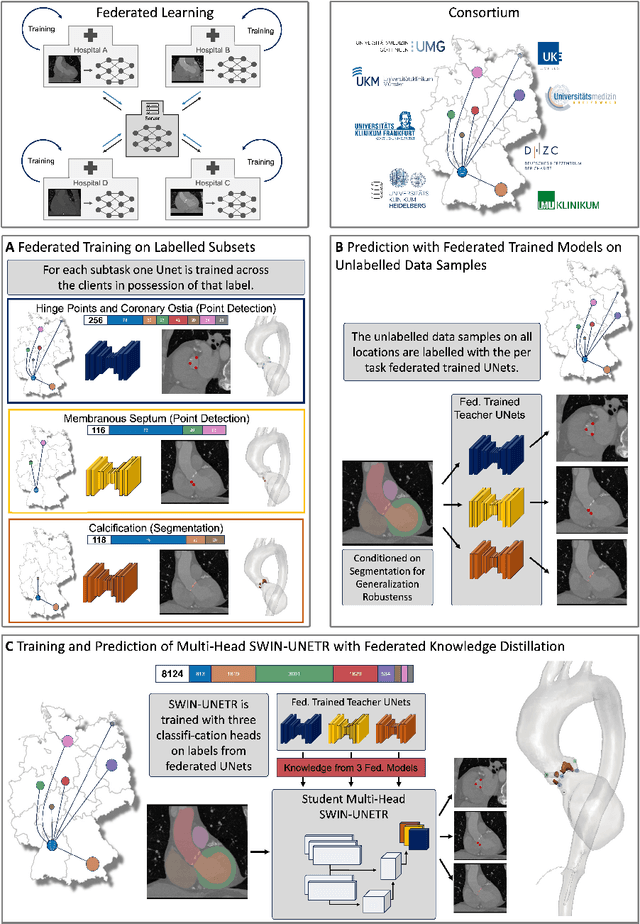
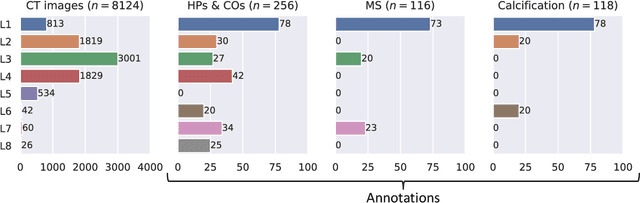
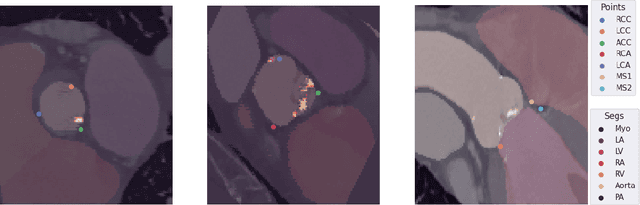

Abstract:Federated learning (FL) is a renowned technique for utilizing decentralized data while preserving privacy. However, real-world applications often involve inherent challenges such as partially labeled datasets, where not all clients possess expert annotations of all labels of interest, leaving large portions of unlabeled data unused. In this study, we conduct the largest federated cardiac CT imaging analysis to date, focusing on partially labeled datasets ($n=8,124$) of Transcatheter Aortic Valve Implantation (TAVI) patients over eight hospital clients. Transformer architectures, which are the major building blocks of current foundation models, have shown superior performance when trained on larger cohorts than traditional CNNs. However, when trained on small task-specific labeled sample sizes, it is currently not feasible to exploit their underlying attention mechanism for improved performance. Therefore, we developed a two-stage semi-supervised learning strategy that distills knowledge from several task-specific CNNs (landmark detection and segmentation of calcification) into a single transformer model by utilizing large amounts of unlabeled data typically residing unused in hospitals to mitigate these issues. This method not only improves the predictive accuracy and generalizability of transformer-based architectures but also facilitates the simultaneous learning of all partial labels within a single transformer model across the federation. Additionally, we show that our transformer-based model extracts more meaningful features for further downstream tasks than the UNet-based one by only training the last layer to also solve segmentation of coronary arteries. We make the code and weights of the final model openly available, which can serve as a foundation model for further research in cardiac CT imaging.
Measuring Orthogonality in Representations of Generative Models
Jul 04, 2024Abstract:In unsupervised representation learning, models aim to distill essential features from high-dimensional data into lower-dimensional learned representations, guided by inductive biases. Understanding the characteristics that make a good representation remains a topic of ongoing research. Disentanglement of independent generative processes has long been credited with producing high-quality representations. However, focusing solely on representations that adhere to the stringent requirements of most disentanglement metrics, may result in overlooking many high-quality representations, well suited for various downstream tasks. These metrics often demand that generative factors be encoded in distinct, single dimensions aligned with the canonical basis of the representation space. Motivated by these observations, we propose two novel metrics: Importance-Weighted Orthogonality (IWO) and Importance-Weighted Rank (IWR). These metrics evaluate the mutual orthogonality and rank of generative factor subspaces. Throughout extensive experiments on common downstream tasks, over several benchmark datasets and models, IWO and IWR consistently show stronger correlations with downstream task performance than traditional disentanglement metrics. Our findings suggest that representation quality is closer related to the orthogonality of independent generative processes rather than their disentanglement, offering a new direction for evaluating and improving unsupervised learning models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge