Malte Tölle
Arbitrary Data as Images: Fusion of Patient Data Across Modalities and Irregular Intervals with Vision Transformers
Jan 30, 2025



Abstract:A patient undergoes multiple examinations in each hospital stay, where each provides different facets of the health status. These assessments include temporal data with varying sampling rates, discrete single-point measurements, therapeutic interventions such as medication administration, and images. While physicians are able to process and integrate diverse modalities intuitively, neural networks need specific modeling for each modality complicating the training procedure. We demonstrate that this complexity can be significantly reduced by visualizing all information as images along with unstructured text and subsequently training a conventional vision-text transformer. Our approach, Vision Transformer for irregular sampled Multi-modal Measurements (ViTiMM), not only simplifies data preprocessing and modeling but also outperforms current state-of-the-art methods in predicting in-hospital mortality and phenotyping, as evaluated on 6,175 patients from the MIMIC-IV dataset. The modalities include patient's clinical measurements, medications, X-ray images, and electrocardiography scans. We hope our work inspires advancements in multi-modal medical AI by reducing the training complexity to (visual) prompt engineering, thus lowering entry barriers and enabling no-code solutions for training. The source code will be made publicly available.
Multi-Modal Dataset Creation for Federated~Learning with DICOM Structured Reports
Jul 12, 2024



Abstract:Purpose: Federated training is often hindered by heterogeneous datasets due to divergent data storage options, inconsistent naming schemes, varied annotation procedures, and disparities in label quality. This is particularly evident in the emerging multi-modal learning paradigms, where dataset harmonization including a uniform data representation and filtering options are of paramount importance. Methods: DICOM structured reports enable the standardized linkage of arbitrary information beyond the imaging domain and can be used within Python deep learning pipelines with highdicom. Building on this, we developed an open platform for data integration and interactive filtering capabilities that simplifies the process of assembling multi-modal datasets. Results: In this study, we extend our prior work by showing its applicability to more and divergent data types, as well as streamlining datasets for federated training within an established consortium of eight university hospitals in Germany. We prove its concurrent filtering ability by creating harmonized multi-modal datasets across all locations for predicting the outcome after minimally invasive heart valve replacement. The data includes DICOM data (i.e. computed tomography images, electrocardiography scans) as well as annotations (i.e. calcification segmentations, pointsets and pacemaker dependency), and metadata (i.e. prosthesis and diagnoses). Conclusion: Structured reports bridge the traditional gap between imaging systems and information systems. Utilizing the inherent DICOM reference system arbitrary data types can be queried concurrently to create meaningful cohorts for clinical studies. The graphical interface as well as example structured report templates will be made publicly available.
Federated Foundation Model for Cardiac CT Imaging
Jul 10, 2024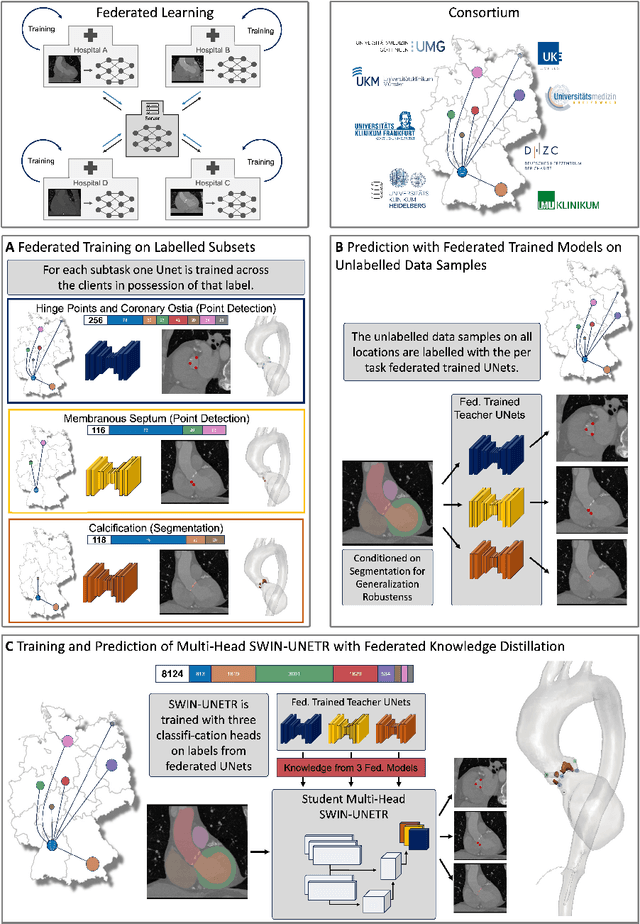
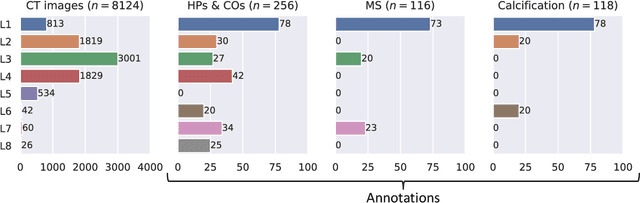
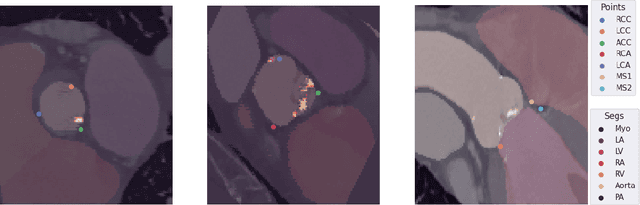

Abstract:Federated learning (FL) is a renowned technique for utilizing decentralized data while preserving privacy. However, real-world applications often involve inherent challenges such as partially labeled datasets, where not all clients possess expert annotations of all labels of interest, leaving large portions of unlabeled data unused. In this study, we conduct the largest federated cardiac CT imaging analysis to date, focusing on partially labeled datasets ($n=8,124$) of Transcatheter Aortic Valve Implantation (TAVI) patients over eight hospital clients. Transformer architectures, which are the major building blocks of current foundation models, have shown superior performance when trained on larger cohorts than traditional CNNs. However, when trained on small task-specific labeled sample sizes, it is currently not feasible to exploit their underlying attention mechanism for improved performance. Therefore, we developed a two-stage semi-supervised learning strategy that distills knowledge from several task-specific CNNs (landmark detection and segmentation of calcification) into a single transformer model by utilizing large amounts of unlabeled data typically residing unused in hospitals to mitigate these issues. This method not only improves the predictive accuracy and generalizability of transformer-based architectures but also facilitates the simultaneous learning of all partial labels within a single transformer model across the federation. Additionally, we show that our transformer-based model extracts more meaningful features for further downstream tasks than the UNet-based one by only training the last layer to also solve segmentation of coronary arteries. We make the code and weights of the final model openly available, which can serve as a foundation model for further research in cardiac CT imaging.
FUNAvg: Federated Uncertainty Weighted Averaging for Datasets with Diverse Labels
Jul 10, 2024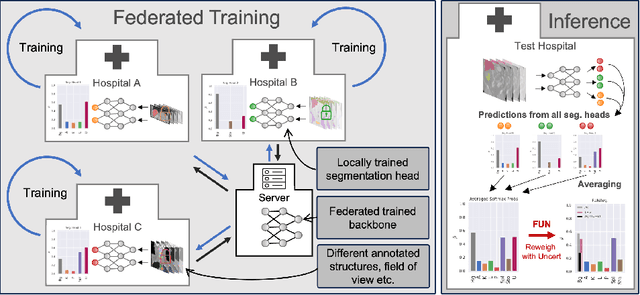

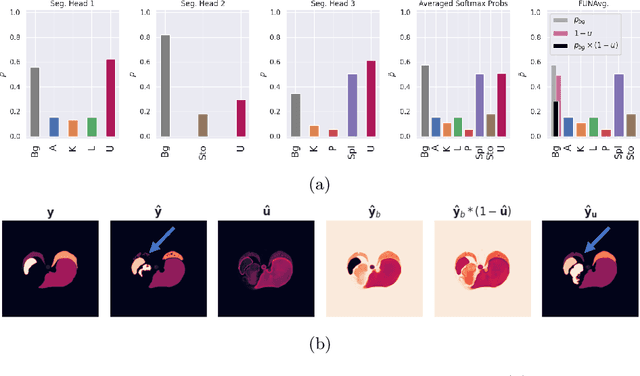

Abstract:Federated learning is one popular paradigm to train a joint model in a distributed, privacy-preserving environment. But partial annotations pose an obstacle meaning that categories of labels are heterogeneous over clients. We propose to learn a joint backbone in a federated manner, while each site receives its own multi-label segmentation head. By using Bayesian techniques we observe that the different segmentation heads although only trained on the individual client's labels also learn information about the other labels not present at the respective site. This information is encoded in their predictive uncertainty. To obtain a final prediction we leverage this uncertainty and perform a weighted averaging of the ensemble of distributed segmentation heads, which allows us to segment "locally unknown" structures. With our method, which we refer to as FUNAvg, we are even on-par with the models trained and tested on the same dataset on average. The code is publicly available at https://github.com/Cardio-AI/FUNAvg.
Content-Aware Differential Privacy with Conditional Invertible Neural Networks
Jul 29, 2022
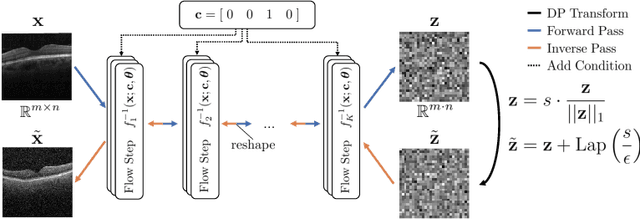
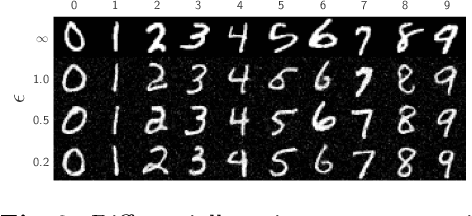
Abstract:Differential privacy (DP) has arisen as the gold standard in protecting an individual's privacy in datasets by adding calibrated noise to each data sample. While the application to categorical data is straightforward, its usability in the context of images has been limited. Contrary to categorical data the meaning of an image is inherent in the spatial correlation of neighboring pixels making the simple application of noise infeasible. Invertible Neural Networks (INN) have shown excellent generative performance while still providing the ability to quantify the exact likelihood. Their principle is based on transforming a complicated distribution into a simple one e.g. an image into a spherical Gaussian. We hypothesize that adding noise to the latent space of an INN can enable differentially private image modification. Manipulation of the latent space leads to a modified image while preserving important details. Further, by conditioning the INN on meta-data provided with the dataset we aim at leaving dimensions important for downstream tasks like classification untouched while altering other parts that potentially contain identifying information. We term our method content-aware differential privacy (CADP). We conduct experiments on publicly available benchmarking datasets as well as dedicated medical ones. In addition, we show the generalizability of our method to categorical data. The source code is publicly available at https://github.com/Cardio-AI/CADP.
Posterior temperature optimized Bayesian models for inverse problems in medical imaging
Feb 02, 2022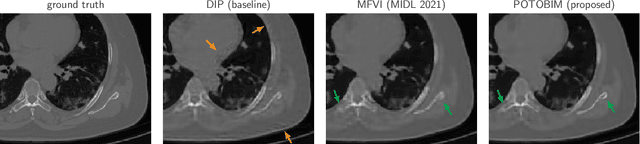
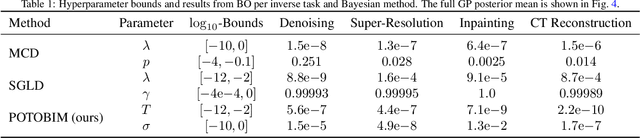
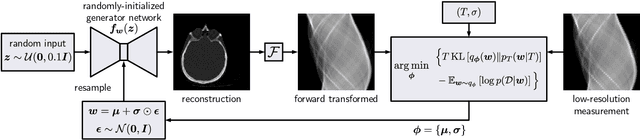
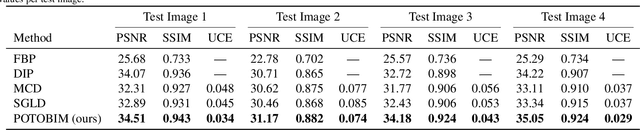
Abstract:We present Posterior Temperature Optimized Bayesian Inverse Models (POTOBIM), an unsupervised Bayesian approach to inverse problems in medical imaging using mean-field variational inference with a fully tempered posterior. Bayesian methods exhibit useful properties for approaching inverse tasks, such as tomographic reconstruction or image denoising. A suitable prior distribution introduces regularization, which is needed to solve the ill-posed problem and reduces overfitting the data. In practice, however, this often results in a suboptimal posterior temperature, and the full potential of the Bayesian approach is not being exploited. In POTOBIM, we optimize both the parameters of the prior distribution and the posterior temperature with respect to reconstruction accuracy using Bayesian optimization with Gaussian process regression. Our method is extensively evaluated on four different inverse tasks on a variety of modalities with images from public data sets and we demonstrate that an optimized posterior temperature outperforms both non-Bayesian and Bayesian approaches without temperature optimization. The use of an optimized prior distribution and posterior temperature leads to improved accuracy and uncertainty estimation and we show that it is sufficient to find these hyperparameters per task domain. Well-tempered posteriors yield calibrated uncertainty, which increases the reliability in the predictions. Our source code is publicly available at github.com/Cardio-AI/mfvi-dip-mia.
Cold Posteriors Improve Bayesian Medical Image Post-Processing
Jul 12, 2021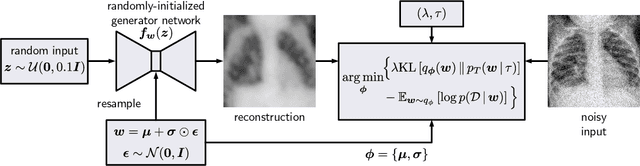
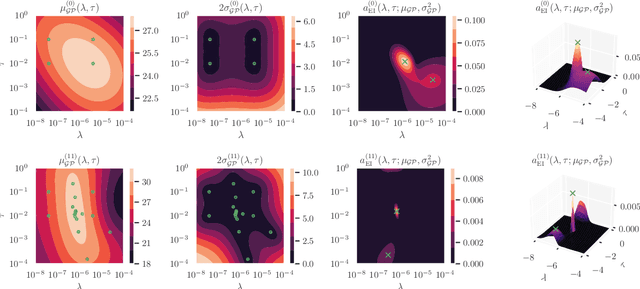
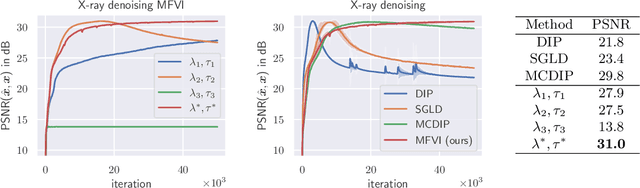
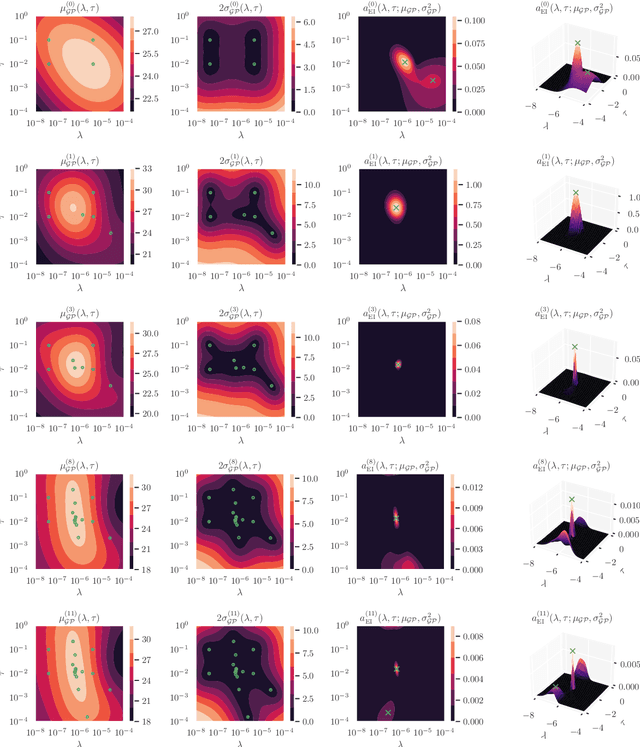
Abstract:Cold posteriors have been reported to perform better in practice in the context of Bayesian deep learning (Wenzel et al., 2020). In variational inference, it is common to employ only a partially tempered posterior by scaling the complexity term in the log-evidence lower bound (ELBO). In this work, we optimize the ELBO for a fully tempered posterior in mean-field variational inference and use Bayesian optimization to automatically find the optimal posterior temperature and prior scale. Choosing an appropriate posterior temperature leads to better predictive performance and improved uncertainty calibration, which we demonstrate for the task of denoising medical X-ray images.
Uncertainty Estimation in Medical Image Denoising with Bayesian Deep Image Prior
Aug 20, 2020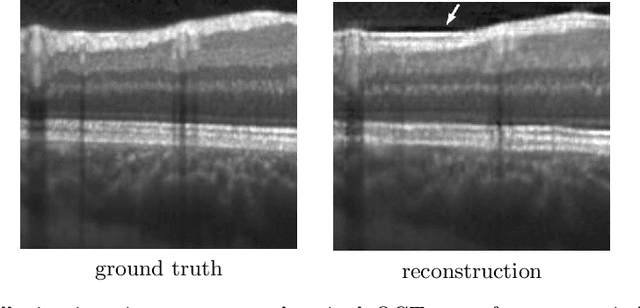
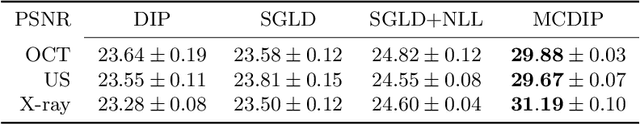
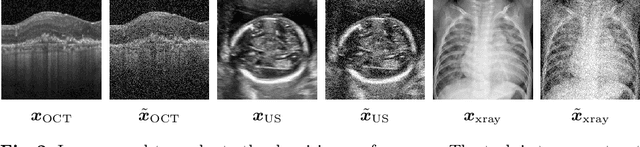

Abstract:Uncertainty quantification in inverse medical imaging tasks with deep learning has received little attention. However, deep models trained on large data sets tend to hallucinate and create artifacts in the reconstructed output that are not anatomically present. We use a randomly initialized convolutional network as parameterization of the reconstructed image and perform gradient descent to match the observation, which is known as deep image prior. In this case, the reconstruction does not suffer from hallucinations as no prior training is performed. We extend this to a Bayesian approach with Monte Carlo dropout to quantify both aleatoric and epistemic uncertainty. The presented method is evaluated on the task of denoising different medical imaging modalities. The experimental results show that our approach yields well-calibrated uncertainty. That is, the predictive uncertainty correlates with the predictive error. This allows for reliable uncertainty estimates and can tackle the problem of hallucinations and artifacts in inverse medical imaging tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge