Halvar Kelm
Multi-Modal Dataset Creation for Federated~Learning with DICOM Structured Reports
Jul 12, 2024



Abstract:Purpose: Federated training is often hindered by heterogeneous datasets due to divergent data storage options, inconsistent naming schemes, varied annotation procedures, and disparities in label quality. This is particularly evident in the emerging multi-modal learning paradigms, where dataset harmonization including a uniform data representation and filtering options are of paramount importance. Methods: DICOM structured reports enable the standardized linkage of arbitrary information beyond the imaging domain and can be used within Python deep learning pipelines with highdicom. Building on this, we developed an open platform for data integration and interactive filtering capabilities that simplifies the process of assembling multi-modal datasets. Results: In this study, we extend our prior work by showing its applicability to more and divergent data types, as well as streamlining datasets for federated training within an established consortium of eight university hospitals in Germany. We prove its concurrent filtering ability by creating harmonized multi-modal datasets across all locations for predicting the outcome after minimally invasive heart valve replacement. The data includes DICOM data (i.e. computed tomography images, electrocardiography scans) as well as annotations (i.e. calcification segmentations, pointsets and pacemaker dependency), and metadata (i.e. prosthesis and diagnoses). Conclusion: Structured reports bridge the traditional gap between imaging systems and information systems. Utilizing the inherent DICOM reference system arbitrary data types can be queried concurrently to create meaningful cohorts for clinical studies. The graphical interface as well as example structured report templates will be made publicly available.
Federated Foundation Model for Cardiac CT Imaging
Jul 10, 2024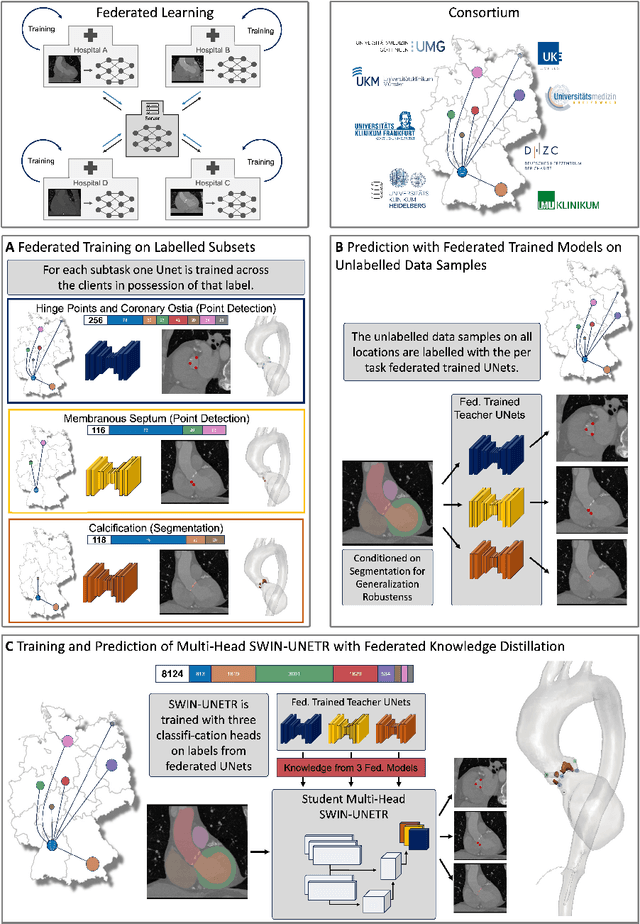
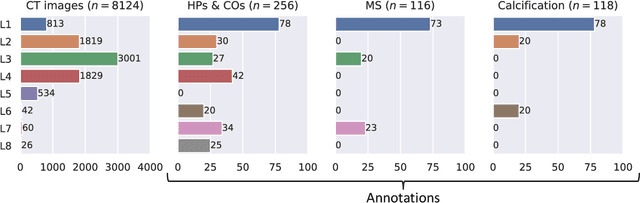
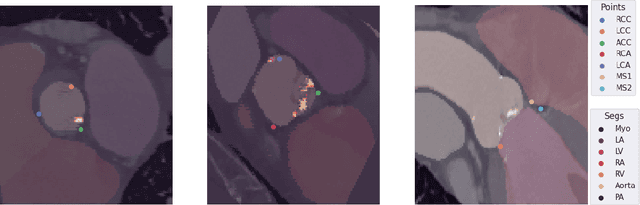

Abstract:Federated learning (FL) is a renowned technique for utilizing decentralized data while preserving privacy. However, real-world applications often involve inherent challenges such as partially labeled datasets, where not all clients possess expert annotations of all labels of interest, leaving large portions of unlabeled data unused. In this study, we conduct the largest federated cardiac CT imaging analysis to date, focusing on partially labeled datasets ($n=8,124$) of Transcatheter Aortic Valve Implantation (TAVI) patients over eight hospital clients. Transformer architectures, which are the major building blocks of current foundation models, have shown superior performance when trained on larger cohorts than traditional CNNs. However, when trained on small task-specific labeled sample sizes, it is currently not feasible to exploit their underlying attention mechanism for improved performance. Therefore, we developed a two-stage semi-supervised learning strategy that distills knowledge from several task-specific CNNs (landmark detection and segmentation of calcification) into a single transformer model by utilizing large amounts of unlabeled data typically residing unused in hospitals to mitigate these issues. This method not only improves the predictive accuracy and generalizability of transformer-based architectures but also facilitates the simultaneous learning of all partial labels within a single transformer model across the federation. Additionally, we show that our transformer-based model extracts more meaningful features for further downstream tasks than the UNet-based one by only training the last layer to also solve segmentation of coronary arteries. We make the code and weights of the final model openly available, which can serve as a foundation model for further research in cardiac CT imaging.
mvHOTA: A multi-view higher order tracking accuracy metric to measure spatial and temporal associations in multi-point detection
Jun 19, 2022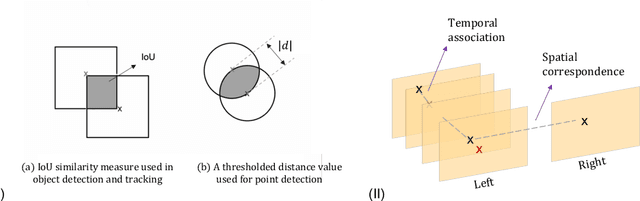
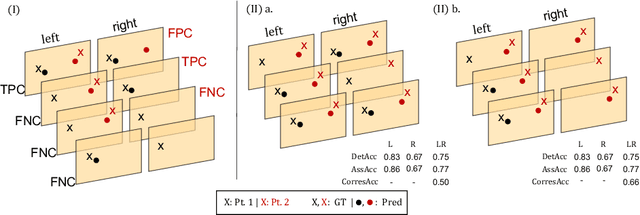
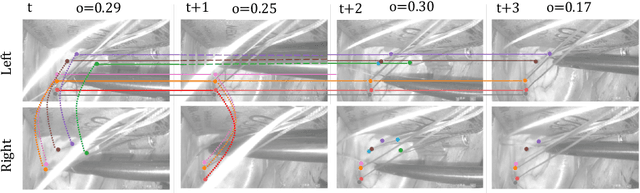
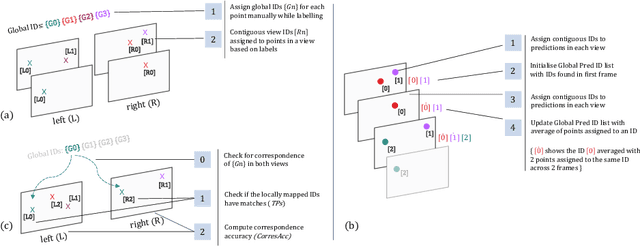
Abstract:Multi-object tracking (MOT) is a challenging task that involves detecting objects in the scene and tracking them across a sequence of frames. Evaluating this task is difficult due to temporal occlusions, and varying trajectories across a sequence of images. The main evaluation metric to benchmark MOT methods on datasets such as KITTI has recently become the higher order tracking accuracy (HOTA) metric, which is capable of providing a better description of the performance over metrics such as MOTA, DetA, and IDF1. Point detection and tracking is a closely related task, which could be regarded as a special case of object detection. However, there are differences in evaluating the detection task itself (point distances vs. bounding box overlap). When including the temporal dimension and multi-view scenarios, the evaluation task becomes even more complex. In this work, we propose a multi-view higher order tracking metric (mvHOTA) to determine the accuracy of multi-point (multi-instance and multi-class) detection, while taking into account temporal and spatial associations. mvHOTA can be interpreted as the geometric mean of the detection, association, and correspondence accuracies, thereby providing equal weighting to each of the factors. We demonstrate a use-case through a publicly available endoscopic point detection dataset from a previously organised medical challenge. Furthermore, we compare with other adjusted MOT metrics for this use-case, discuss the properties of mvHOTA, and show how the proposed correspondence accuracy and the Occlusion index facilitate analysis of methods with respect to handling of occlusions. The code will be made publicly available.
Point detection through multi-instance deep heatmap regression for sutures in endoscopy
Nov 16, 2021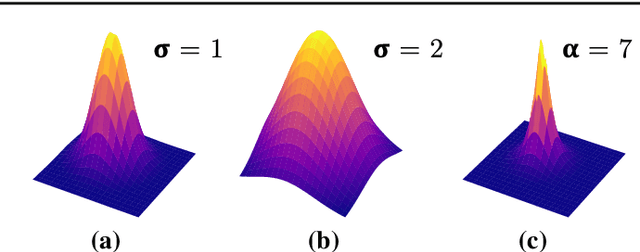
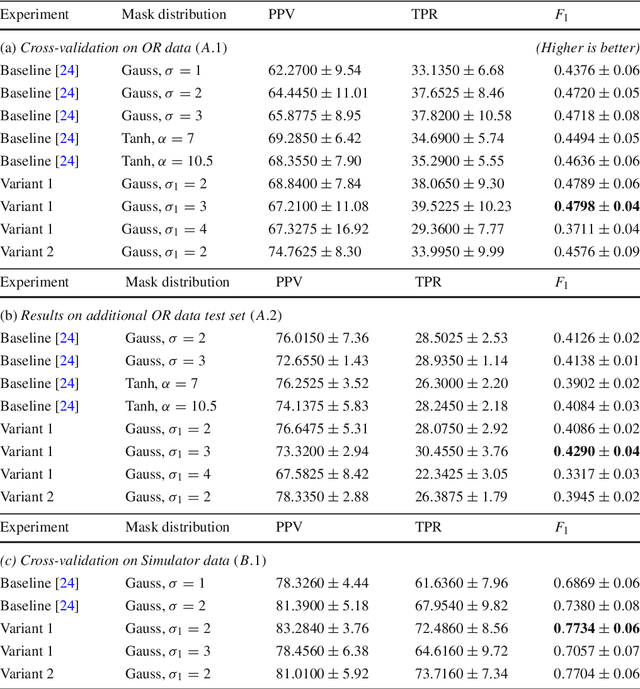
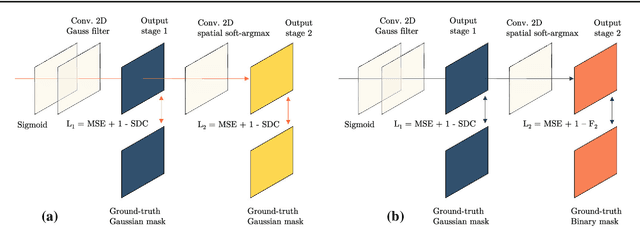
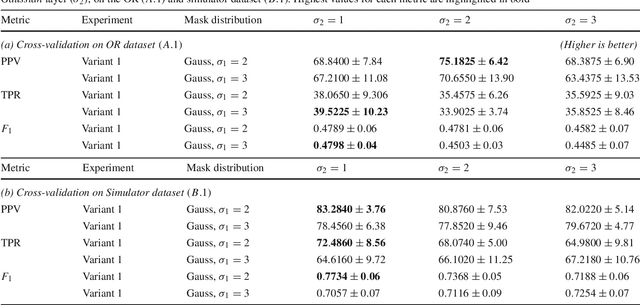
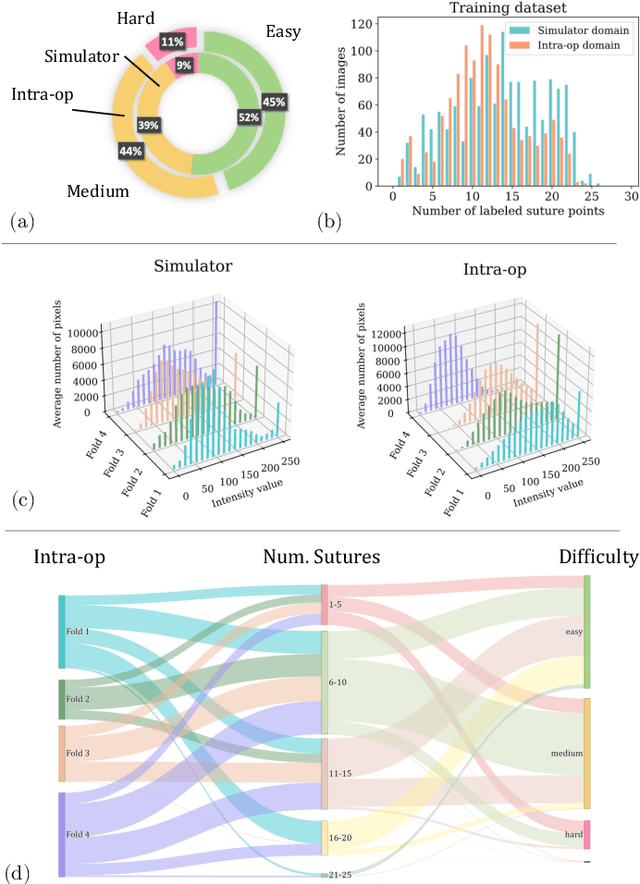
Abstract:Purpose: Mitral valve repair is a complex minimally invasive surgery of the heart valve. In this context, suture detection from endoscopic images is a highly relevant task that provides quantitative information to analyse suturing patterns, assess prosthetic configurations and produce augmented reality visualisations. Facial or anatomical landmark detection tasks typically contain a fixed number of landmarks, and use regression or fixed heatmap-based approaches to localize the landmarks. However in endoscopy, there are a varying number of sutures in every image, and the sutures may occur at any location in the annulus, as they are not semantically unique. Method: In this work, we formulate the suture detection task as a multi-instance deep heatmap regression problem, to identify entry and exit points of sutures. We extend our previous work, and introduce the novel use of a 2D Gaussian layer followed by a differentiable 2D spatial Soft-Argmax layer to function as a local non-maximum suppression. Results: We present extensive experiments with multiple heatmap distribution functions and two variants of the proposed model. In the intra-operative domain, Variant 1 showed a mean F1 of +0.0422 over the baseline. Similarly, in the simulator domain, Variant 1 showed a mean F1 of +0.0865 over the baseline. Conclusion: The proposed model shows an improvement over the baseline in the intra-operative and the simulator domains. The data is made publicly available within the scope of the MICCAI AdaptOR2021 Challenge https://adaptor2021.github.io/, and the code at https://github.com/Cardio-AI/suture-detection-pytorch/. DOI:10.1007/s11548-021-02523-w. The link to the open access article can be found here: https://link.springer.com/article/10.1007%2Fs11548-021-02523-w
* Accepted to International Journal of Computer Assisted Radiology and Surgery, 15 pages, 5 figures
Mutually improved endoscopic image synthesis and landmark detection in unpaired image-to-image translation
Jul 14, 2021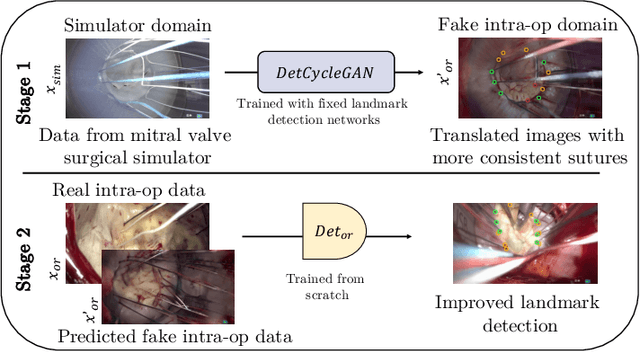
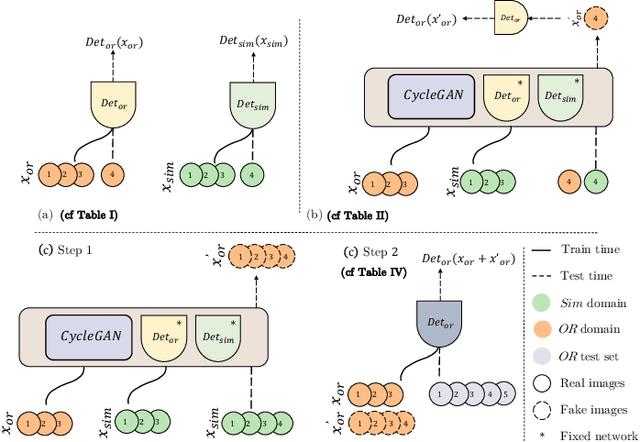
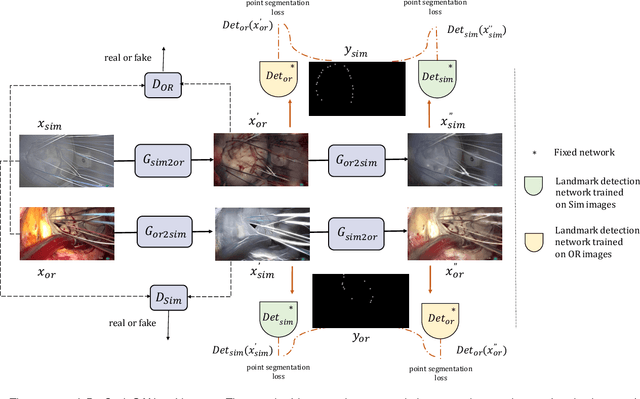
Abstract:The CycleGAN framework allows for unsupervised image-to-image translation of unpaired data. In a scenario of surgical training on a physical surgical simulator, this method can be used to transform endoscopic images of phantoms into images which more closely resemble the intra-operative appearance of the same surgical target structure. This can be viewed as a novel augmented reality approach, which we coined Hyperrealism in previous work. In this use case, it is of paramount importance to display objects like needles, sutures or instruments consistent in both domains while altering the style to a more tissue-like appearance. Segmentation of these objects would allow for a direct transfer, however, contouring of these, partly tiny and thin foreground objects is cumbersome and perhaps inaccurate. Instead, we propose to use landmark detection on the points when sutures pass into the tissue. This objective is directly incorporated into a CycleGAN framework by treating the performance of pre-trained detector models as an additional optimization goal. We show that a task defined on these sparse landmark labels improves consistency of synthesis by the generator network in both domains. Comparing a baseline CycleGAN architecture to our proposed extension (DetCycleGAN), mean precision (PPV) improved by +61.32, mean sensitivity (TPR) by +37.91, and mean F1 score by +0.4743. Furthermore, it could be shown that by dataset fusion, generated intra-operative images can be leveraged as additional training data for the detection network itself. The data is released within the scope of the AdaptOR MICCAI Challenge 2021 at https://adaptor2021.github.io/, and code at https://github.com/Cardio-AI/detcyclegan_pytorch.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge