Anja Hennemuth
Carotid Artery Plaque Analysis in 3D Based on Distance Encoding in Mesh Representations
Feb 18, 2025Abstract:Purpose: Enabling a comprehensive and robust assessment of carotid artery plaques in 3D through extraction and visualization of quantitative plaque parameters. These parameters have potential applications in stroke risk analysis, evaluation of therapy effectiveness, and plaque progression prediction. Methods: We propose a novel method for extracting a plaque mesh from 3D vessel wall segmentation using distance encoding on the inner and outer wall mesh for precise plaque structure analysis. A case-specific threshold, derived from the normal vessel wall thickness, was applied to extract plaques from a dataset of 202 T1-weighted black-blood MRI scans of subjects with up to 50% stenosis. Applied to baseline and one-year follow-up data, the method supports detailed plaque morphology analysis over time, including plaque volume quantification, aided by improved visualization via mesh unfolding. Results: We successfully extracted plaque meshes from 341 carotid arteries, capturing a wide range of plaque shapes with volumes ranging from 2.69{\mu}l to 847.7{\mu}l. The use of a case-specific threshold effectively eliminated false positives in young, healthy subjects. Conclusion: The proposed method enables precise extraction of plaque meshes from 3D vessel wall segmentation masks enabling a correspondence between baseline and one-year follow-up examinations. Unfolding the plaque meshes enhances visualization, while the mesh-based analysis allows quantification of plaque parameters independent of voxel resolution.
Learning Wall Segmentation in 3D Vessel Trees using Sparse Annotations
Feb 18, 2025Abstract:We propose a novel approach that uses sparse annotations from clinical studies to train a 3D segmentation of the carotid artery wall. We use a centerline annotation to sample perpendicular cross-sections of the carotid artery and use an adversarial 2D network to segment them. These annotations are then transformed into 3D pseudo-labels for training of a 3D convolutional neural network, circumventing the creation of manual 3D masks. For pseudo-label creation in the bifurcation area we propose the use of cross-sections perpendicular to the bifurcation axis and show that this enhances segmentation performance. Different sampling distances had a lesser impact. The proposed method allows for efficient training of 3D segmentation, offering potential improvements in the assessment of carotid artery stenosis and allowing the extraction of 3D biomarkers such as plaque volume.
Multi-Modal Dataset Creation for Federated~Learning with DICOM Structured Reports
Jul 12, 2024



Abstract:Purpose: Federated training is often hindered by heterogeneous datasets due to divergent data storage options, inconsistent naming schemes, varied annotation procedures, and disparities in label quality. This is particularly evident in the emerging multi-modal learning paradigms, where dataset harmonization including a uniform data representation and filtering options are of paramount importance. Methods: DICOM structured reports enable the standardized linkage of arbitrary information beyond the imaging domain and can be used within Python deep learning pipelines with highdicom. Building on this, we developed an open platform for data integration and interactive filtering capabilities that simplifies the process of assembling multi-modal datasets. Results: In this study, we extend our prior work by showing its applicability to more and divergent data types, as well as streamlining datasets for federated training within an established consortium of eight university hospitals in Germany. We prove its concurrent filtering ability by creating harmonized multi-modal datasets across all locations for predicting the outcome after minimally invasive heart valve replacement. The data includes DICOM data (i.e. computed tomography images, electrocardiography scans) as well as annotations (i.e. calcification segmentations, pointsets and pacemaker dependency), and metadata (i.e. prosthesis and diagnoses). Conclusion: Structured reports bridge the traditional gap between imaging systems and information systems. Utilizing the inherent DICOM reference system arbitrary data types can be queried concurrently to create meaningful cohorts for clinical studies. The graphical interface as well as example structured report templates will be made publicly available.
Federated Foundation Model for Cardiac CT Imaging
Jul 10, 2024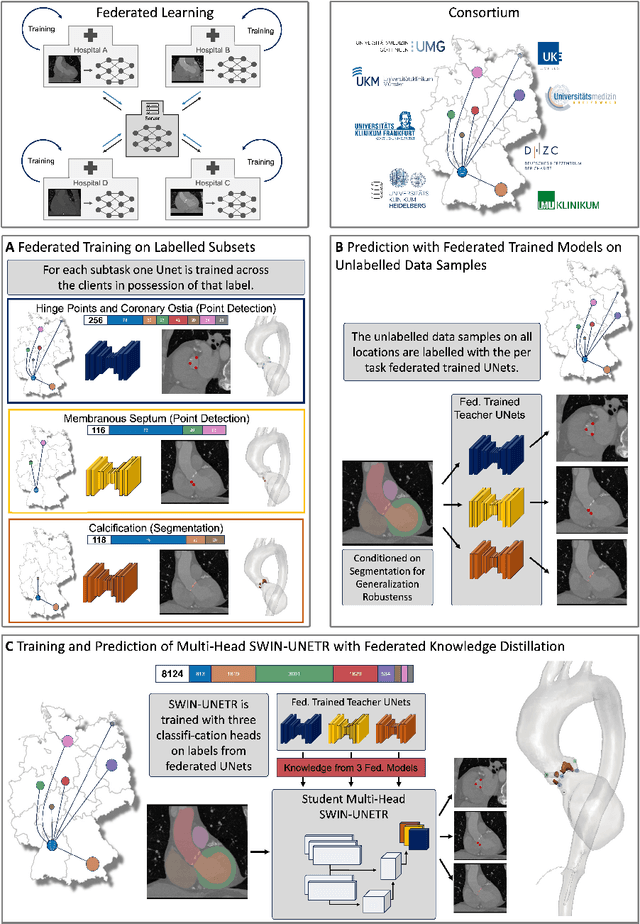
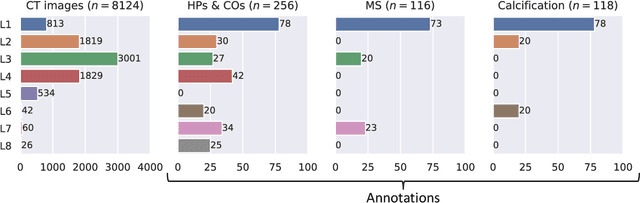
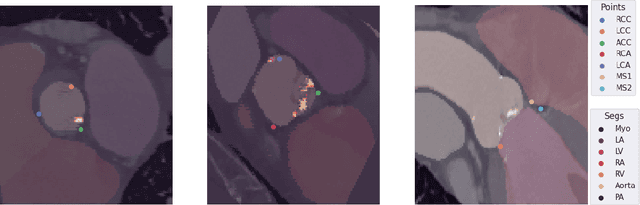

Abstract:Federated learning (FL) is a renowned technique for utilizing decentralized data while preserving privacy. However, real-world applications often involve inherent challenges such as partially labeled datasets, where not all clients possess expert annotations of all labels of interest, leaving large portions of unlabeled data unused. In this study, we conduct the largest federated cardiac CT imaging analysis to date, focusing on partially labeled datasets ($n=8,124$) of Transcatheter Aortic Valve Implantation (TAVI) patients over eight hospital clients. Transformer architectures, which are the major building blocks of current foundation models, have shown superior performance when trained on larger cohorts than traditional CNNs. However, when trained on small task-specific labeled sample sizes, it is currently not feasible to exploit their underlying attention mechanism for improved performance. Therefore, we developed a two-stage semi-supervised learning strategy that distills knowledge from several task-specific CNNs (landmark detection and segmentation of calcification) into a single transformer model by utilizing large amounts of unlabeled data typically residing unused in hospitals to mitigate these issues. This method not only improves the predictive accuracy and generalizability of transformer-based architectures but also facilitates the simultaneous learning of all partial labels within a single transformer model across the federation. Additionally, we show that our transformer-based model extracts more meaningful features for further downstream tasks than the UNet-based one by only training the last layer to also solve segmentation of coronary arteries. We make the code and weights of the final model openly available, which can serve as a foundation model for further research in cardiac CT imaging.
Deep Learning methods for automatic evaluation of delayed enhancement-MRI. The results of the EMIDEC challenge
Aug 10, 2021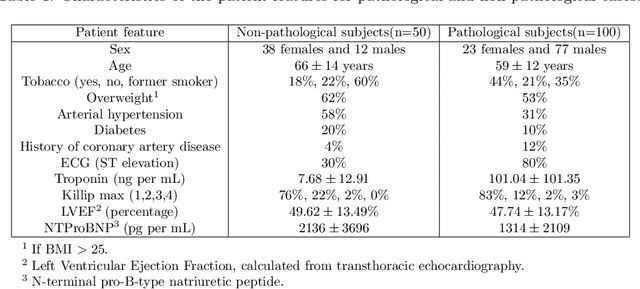
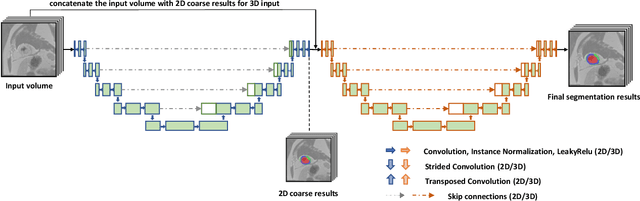
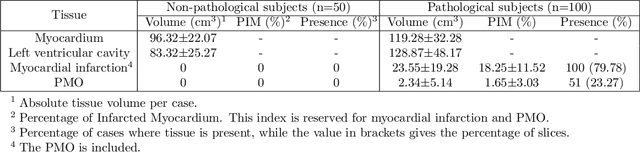
Abstract:A key factor for assessing the state of the heart after myocardial infarction (MI) is to measure whether the myocardium segment is viable after reperfusion or revascularization therapy. Delayed enhancement-MRI or DE-MRI, which is performed several minutes after injection of the contrast agent, provides high contrast between viable and nonviable myocardium and is therefore a method of choice to evaluate the extent of MI. To automatically assess myocardial status, the results of the EMIDEC challenge that focused on this task are presented in this paper. The challenge's main objectives were twofold. First, to evaluate if deep learning methods can distinguish between normal and pathological cases. Second, to automatically calculate the extent of myocardial infarction. The publicly available database consists of 150 exams divided into 50 cases with normal MRI after injection of a contrast agent and 100 cases with myocardial infarction (and then with a hyperenhanced area on DE-MRI), whatever their inclusion in the cardiac emergency department. Along with MRI, clinical characteristics are also provided. The obtained results issued from several works show that the automatic classification of an exam is a reachable task (the best method providing an accuracy of 0.92), and the automatic segmentation of the myocardium is possible. However, the segmentation of the diseased area needs to be improved, mainly due to the small size of these areas and the lack of contrast with the surrounding structures.
Fully automated quantification of in vivo viscoelasticity of prostate zones using magnetic resonance elastography with Dense U-net segmentation
Jun 21, 2021



Abstract:Magnetic resonance elastography (MRE) for measuring viscoelasticity heavily depends on proper tissue segmentation, especially in heterogeneous organs such as the prostate. Using trained network-based image segmentation, we investigated if MRE data suffice to extract anatomical and viscoelastic information for automatic tabulation of zonal mechanical properties of the prostate. Overall, 40 patients with benign prostatic hyperplasia (BPH) or prostate cancer (PCa) were examined with three magnetic resonance imaging (MRI) sequences: T2-weighted MRI (T2w), diffusion-weighted imaging (DWI), and MRE-based tomoelastography yielding six independent sets of imaging data per patient (T2w, DWI, apparent diffusion coefficient (ADC), MRE magnitude, shear wave speed, and loss angle maps). Combinations of these data were used to train Dense U-nets with manually segmented masks of the entire prostate gland (PG), central zone (CZ), and peripheral zone (PZ) in 30 patients and to validate them in 10 patients. Dice score (DS), sensitivity, specificity, and Hausdorff distance were determined. We found that segmentation based on MRE magnitude maps alone (DS, PG: 0.93$\pm$0.04, CZ: 0.95$\pm$0.03, PZ: 0.77$\pm$0.05) was more accurate than magnitude maps combined with T2w and DWI_b (DS, PG: 0.91$\pm$0.04, CZ: 0.91$\pm$0.06, PZ: 0.63$\pm$0.16) or T2w alone (DS, PG: 0.92$\pm$0.03, CZ: 0.91$\pm$0.04, PZ: 0.65$\pm$0.08). Automatically tabulated MRE values were not different from ground-truth values (P>0.05). In conclusion: MRE combined with Dense U-net segmentation allows tabulation of quantitative imaging markers without manual analysis and independent of other MRI sequences and can thus contribute to PCa detection and classification.
Anonymization of labeled TOF-MRA images for brain vessel segmentation using generative adversarial networks
Sep 16, 2020



Abstract:Anonymization and data sharing are crucial for privacy protection and acquisition of large datasets for medical image analysis. This is a big challenge, especially for neuroimaging. Here, the brain's unique structure allows for re-identification and thus requires non-conventional anonymization. Generative adversarial networks (GANs) have the potential to provide anonymous images while preserving predictive properties. Analyzing brain vessel segmentation as a use case, we trained 3 GANs on time-of-flight (TOF) magnetic resonance angiography (MRA) patches for image-label generation: 1) Deep convolutional GAN, 2) Wasserstein-GAN with gradient penalty (WGAN-GP) and 3) WGAN-GP with spectral normalization (WGAN-GP-SN). The generated image-labels from each GAN were used to train a U-net for segmentation and tested on real data. Moreover, we applied our synthetic patches using transfer learning on a second dataset. For an increasing number of up to 15 patients we evaluated the model performance on real data with and without pre-training. The performance for all models was assessed by the Dice Similarity Coefficient (DSC) and the 95th percentile of the Hausdorff Distance (95HD). Comparing the 3 GANs, the U-net trained on synthetic data generated by the WGAN-GP-SN showed the highest performance to predict vessels (DSC/95HD 0.82/28.97) benchmarked by the U-net trained on real data (0.89/26.61). The transfer learning approach showed superior performance for the same GAN compared to no pre-training, especially for one patient only (0.91/25.68 vs. 0.85/27.36). In this work, synthetic image-label pairs retained generalizable information and showed good performance for vessel segmentation. Besides, we showed that synthetic patches can be used in a transfer learning approach with independent data. This paves the way to overcome the challenges of scarce data and anonymization in medical imaging.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge