Sebastian Schmitter
Carotid Artery Plaque Analysis in 3D Based on Distance Encoding in Mesh Representations
Feb 18, 2025Abstract:Purpose: Enabling a comprehensive and robust assessment of carotid artery plaques in 3D through extraction and visualization of quantitative plaque parameters. These parameters have potential applications in stroke risk analysis, evaluation of therapy effectiveness, and plaque progression prediction. Methods: We propose a novel method for extracting a plaque mesh from 3D vessel wall segmentation using distance encoding on the inner and outer wall mesh for precise plaque structure analysis. A case-specific threshold, derived from the normal vessel wall thickness, was applied to extract plaques from a dataset of 202 T1-weighted black-blood MRI scans of subjects with up to 50% stenosis. Applied to baseline and one-year follow-up data, the method supports detailed plaque morphology analysis over time, including plaque volume quantification, aided by improved visualization via mesh unfolding. Results: We successfully extracted plaque meshes from 341 carotid arteries, capturing a wide range of plaque shapes with volumes ranging from 2.69{\mu}l to 847.7{\mu}l. The use of a case-specific threshold effectively eliminated false positives in young, healthy subjects. Conclusion: The proposed method enables precise extraction of plaque meshes from 3D vessel wall segmentation masks enabling a correspondence between baseline and one-year follow-up examinations. Unfolding the plaque meshes enhances visualization, while the mesh-based analysis allows quantification of plaque parameters independent of voxel resolution.
Learning Wall Segmentation in 3D Vessel Trees using Sparse Annotations
Feb 18, 2025Abstract:We propose a novel approach that uses sparse annotations from clinical studies to train a 3D segmentation of the carotid artery wall. We use a centerline annotation to sample perpendicular cross-sections of the carotid artery and use an adversarial 2D network to segment them. These annotations are then transformed into 3D pseudo-labels for training of a 3D convolutional neural network, circumventing the creation of manual 3D masks. For pseudo-label creation in the bifurcation area we propose the use of cross-sections perpendicular to the bifurcation axis and show that this enhances segmentation performance. Different sampling distances had a lesser impact. The proposed method allows for efficient training of 3D segmentation, offering potential improvements in the assessment of carotid artery stenosis and allowing the extraction of 3D biomarkers such as plaque volume.
DeepControl: 2D RF pulses facilitating $B_1^+$ inhomogeneity and $B_0$ off-resonance compensation in vivo at 7T
Sep 25, 2020
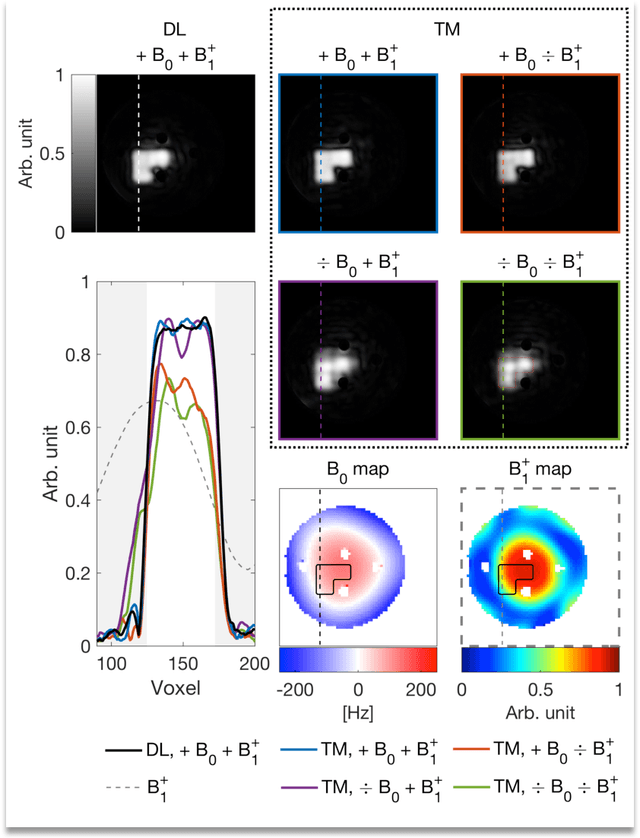
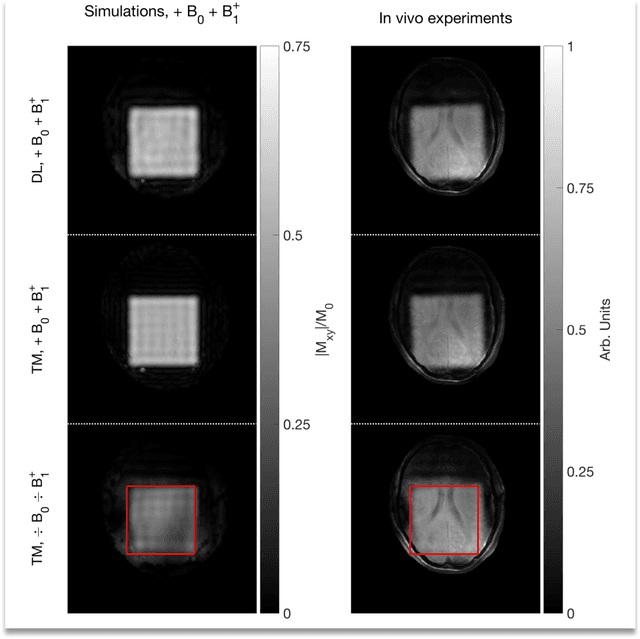

Abstract:Purpose: Rapid 2D RF pulse design with subject specific $B_1^+$ inhomogeneity and $B_0$ off-resonance compensation at 7 T predicted from convolutional neural networks is presented. Methods: The convolution neural network was trained on half a million single-channel transmit, 2D RF pulses optimized with an optimal control method using artificial 2D targets, $B_1^+$ and $B_0$ maps. Predicted pulses were tested in a phantom and in vivo at 7 T with measured $B_1^+$ and $B_0$ maps from a high-resolution GRE sequence. Results: Pulse prediction by the trained convolutional neural network was done on the fly during the MR session in approximately 9 ms for multiple hand drawn ROIs and the measured $B_1^+$ and $B_0$ maps. Compensation of $B_1^+$ inhomogeneity and $B_0$ off-resonances has been confirmed in the phantom and in vivo experiments. The reconstructed image data agrees well with the simulations using the acquired $B_1^+$ and $B_0$ maps and the 2D RF pulse predicted by the convolutional neural networks is as good as the conventional RF pulse obtained by optimal control. Conclusion: The proposed convolutional neural network based 2D RF pulse design method predicts 2D RF pulses with an excellent excitation pattern and compensated $B_1^+$ and $B_0$ variations at 7 T. The rapid 2D RF pulse prediction (9 ms) enables subject-specific high-quality 2D RF pulses without the need to run lengthy optimizations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge