Ivo Wolf
FUNAvg: Federated Uncertainty Weighted Averaging for Datasets with Diverse Labels
Jul 10, 2024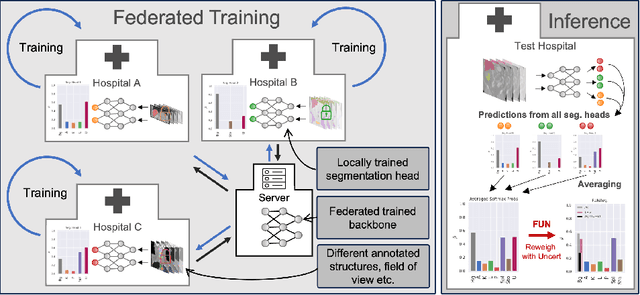

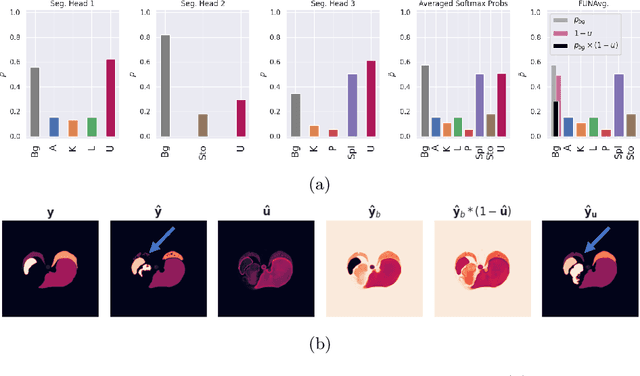

Abstract:Federated learning is one popular paradigm to train a joint model in a distributed, privacy-preserving environment. But partial annotations pose an obstacle meaning that categories of labels are heterogeneous over clients. We propose to learn a joint backbone in a federated manner, while each site receives its own multi-label segmentation head. By using Bayesian techniques we observe that the different segmentation heads although only trained on the individual client's labels also learn information about the other labels not present at the respective site. This information is encoded in their predictive uncertainty. To obtain a final prediction we leverage this uncertainty and perform a weighted averaging of the ensemble of distributed segmentation heads, which allows us to segment "locally unknown" structures. With our method, which we refer to as FUNAvg, we are even on-par with the models trained and tested on the same dataset on average. The code is publicly available at https://github.com/Cardio-AI/FUNAvg.
Anatomy-informed Data Augmentation for Enhanced Prostate Cancer Detection
Sep 07, 2023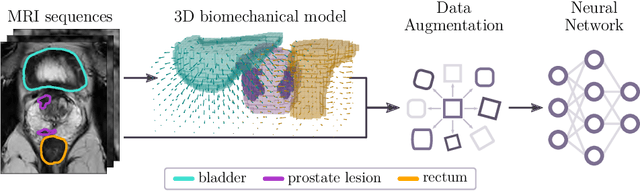

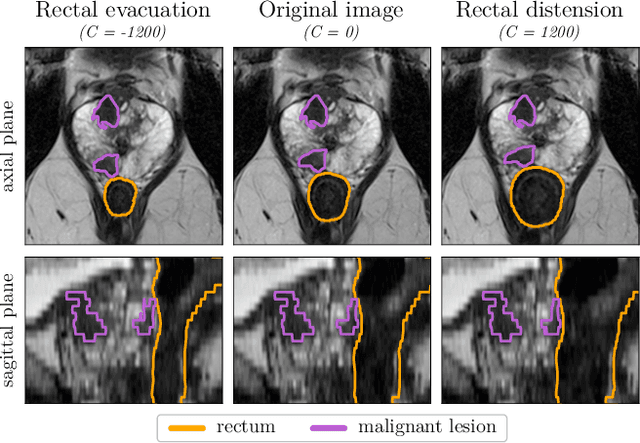
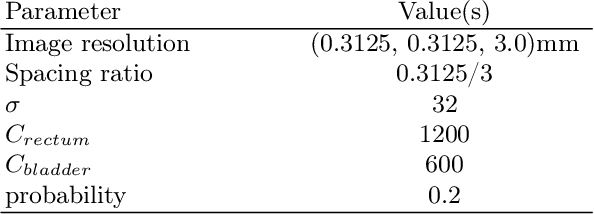
Abstract:Data augmentation (DA) is a key factor in medical image analysis, such as in prostate cancer (PCa) detection on magnetic resonance images. State-of-the-art computer-aided diagnosis systems still rely on simplistic spatial transformations to preserve the pathological label post transformation. However, such augmentations do not substantially increase the organ as well as tumor shape variability in the training set, limiting the model's ability to generalize to unseen cases with more diverse localized soft-tissue deformations. We propose a new anatomy-informed transformation that leverages information from adjacent organs to simulate typical physiological deformations of the prostate and generates unique lesion shapes without altering their label. Due to its lightweight computational requirements, it can be easily integrated into common DA frameworks. We demonstrate the effectiveness of our augmentation on a dataset of 774 biopsy-confirmed examinations, by evaluating a state-of-the-art method for PCa detection with different augmentation settings.
Surgical Phase and Instrument Recognition: How to identify appropriate Dataset Splits
Jun 29, 2023Abstract:Purpose: The development of machine learning models for surgical workflow and instrument recognition from temporal data represents a challenging task due to the complex nature of surgical workflows. In particular, the imbalanced distribution of data is one of the major challenges in the domain of surgical workflow recognition. In order to obtain meaningful results, careful partitioning of data into training, validation, and test sets, as well as the selection of suitable evaluation metrics are crucial. Methods: In this work, we present an openly available web-based application that enables interactive exploration of dataset partitions. The proposed visual framework facilitates the assessment of dataset splits for surgical workflow recognition, especially with regard to identifying sub-optimal dataset splits. Currently, it supports visualization of surgical phase and instrument annotations. Results: In order to validate the dedicated interactive visualizations, we use a dataset split of the Cholec80 dataset. This dataset split was specifically selected to reflect a case of strong data imbalance. Using our software, we were able to identify phases, phase transitions, and combinations of surgical instruments that were not represented in one of the sets. Conclusion: In order to obtain meaningful results in highly unbalanced class distributions, special care should be taken with respect to the selection of an appropriate split. Interactive data visualization represents a promising approach for the assessment of machine learning datasets. The source code is available at https://github.com/Cardio-AI/endovis-ml
Comparison of Depth Estimation Setups from Stereo Endoscopy and Optical Tracking for Point Measurements
Jan 26, 2022

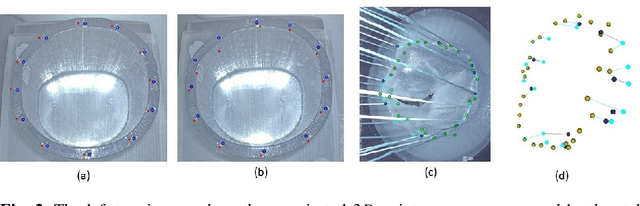
Abstract:To support minimally-invasive intraoperative mitral valve repair, quantitative measurements from the valve can be obtained using an infra-red tracked stylus. It is desirable to view such manually measured points together with the endoscopic image for further assistance. Therefore, hand-eye calibration is required that links both coordinate systems and is a prerequisite to project the points onto the image plane. A complementary approach to this is to use a vision-based endoscopic stereo-setup to detect and triangulate points of interest, to obtain the 3D coordinates. In this paper, we aim to compare both approaches on a rigid phantom and two patient-individual silicone replica which resemble the intraoperative scenario. The preliminary results indicate that 3D landmark estimation, either labeled manually or through partly automated detection with a deep learning approach, provides more accurate triangulated depth measurements when performed with a tailored image-based method than with stylus measurements.
Unsupervised Domain Adaptation from Axial to Short-Axis Multi-Slice Cardiac MR Images by Incorporating Pretrained Task Networks
Jan 20, 2021



Abstract:Anisotropic multi-slice Cardiac Magnetic Resonance (CMR) Images are conventionally acquired in patient-specific short-axis (SAX) orientation. In specific cardiovascular diseases that affect right ventricular (RV) morphology, acquisitions in standard axial (AX) orientation are preferred by some investigators, due to potential superiority in RV volume measurement for treatment planning. Unfortunately, due to the rare occurrence of these diseases, data in this domain is scarce. Recent research in deep learning-based methods mainly focused on SAX CMR images and they had proven to be very successful. In this work, we show that there is a considerable domain shift between AX and SAX images, and therefore, direct application of existing models yield sub-optimal results on AX samples. We propose a novel unsupervised domain adaptation approach, which uses task-related probabilities in an attention mechanism. Beyond that, cycle consistency is imposed on the learned patient-individual 3D rigid transformation to improve stability when automatically re-sampling the AX images to SAX orientations. The network was trained on 122 registered 3D AX-SAX CMR volume pairs from a multi-centric patient cohort. A mean 3D Dice of $0.86\pm{0.06}$ for the left ventricle, $0.65\pm{0.08}$ for the myocardium, and $0.77\pm{0.10}$ for the right ventricle could be achieved. This is an improvement of $25\%$ in Dice for RV in comparison to direct application on axial slices. To conclude, our pre-trained task module has neither seen CMR images nor labels from the target domain, but is able to segment them after the domain gap is reduced. Code: https://github.com/Cardio-AI/3d-mri-domain-adaptation
Heatmap-based 2D Landmark Detection with a Varying Number of Landmarks
Jan 07, 2021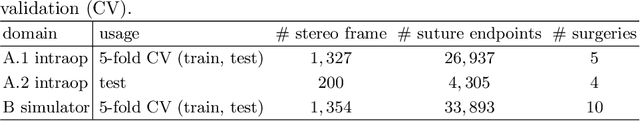
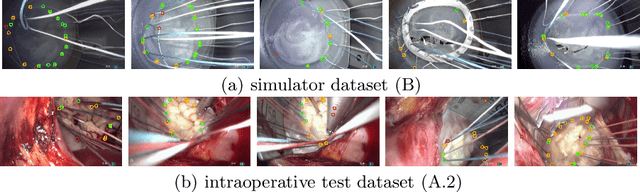
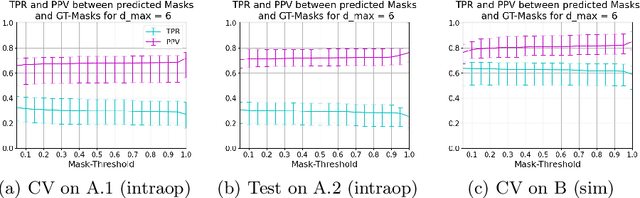
Abstract:Mitral valve repair is a surgery to restore the function of the mitral valve. To achieve this, a prosthetic ring is sewed onto the mitral annulus. Analyzing the sutures, which are punctured through the annulus for ring implantation, can be useful in surgical skill assessment, for quantitative surgery and for positioning a virtual prosthetic ring model in the scene via augmented reality. This work presents a neural network approach which detects the sutures in endoscopic images of mitral valve repair and therefore solves a landmark detection problem with varying amount of landmarks, as opposed to most other existing deep learning-based landmark detection approaches. The neural network is trained separately on two data collections from different domains with the same architecture and hyperparameter settings. The datasets consist of more than 1,300 stereo frame pairs each, with a total over 60,000 annotated landmarks. The proposed heatmap-based neural network achieves a mean positive predictive value (PPV) of 66.68$\pm$4.67% and a mean true positive rate (TPR) of 24.45$\pm$5.06% on the intraoperative test dataset and a mean PPV of 81.50\pm5.77\% and a mean TPR of 61.60$\pm$6.11% on a dataset recorded during surgical simulation. The best detection results are achieved when the camera is positioned above the mitral valve with good illumination. A detection from a sideward view is also possible if the mitral valve is well perceptible.
How well do U-Net-based segmentation trained on adult cardiac magnetic resonance imaging data generalise to rare congenital heart diseases for surgical planning?
Feb 10, 2020



Abstract:Planning the optimal time of intervention for pulmonary valve replacement surgery in patients with the congenital heart disease Tetralogy of Fallot (TOF) is mainly based on ventricular volume and function according to current guidelines. Both of these two biomarkers are most reliably assessed by segmentation of 3D cardiac magnetic resonance (CMR) images. In several grand challenges in the last years, U-Net architectures have shown impressive results on the provided data. However, in clinical practice, data sets are more diverse considering individual pathologies and image properties derived from different scanner properties. Additionally, specific training data for complex rare diseases like TOF is scarce. For this work, 1) we assessed the accuracy gap when using a publicly available labelled data set (the Automatic Cardiac Diagnosis Challenge (ACDC) data set) for training and subsequent applying it to CMR data of TOF patients and vice versa and 2) whether we can achieve similar results when applying the model to a more heterogeneous data base. Multiple deep learning models were trained with four-fold cross validation. Afterwards they were evaluated on the respective unseen CMR images from the other collection. Our results confirm that current deep learning models can achieve excellent results (left ventricle dice of $0.951\pm{0.003}$/$0.941\pm{0.007}$ train/validation) within a single data collection. But once they are applied to other pathologies, it becomes apparent how much they overfit to the training pathologies (dice score drops between $0.072\pm{0.001}$ for the left and $0.165\pm{0.001}$ for the right ventricle).
Cross-Domain Conditional Generative Adversarial Networks for Stereoscopic Hyperrealism in Surgical Training
Jun 24, 2019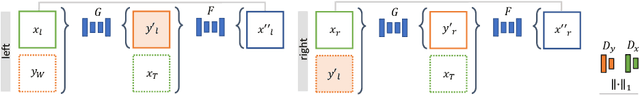
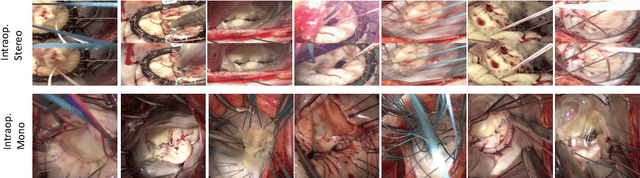
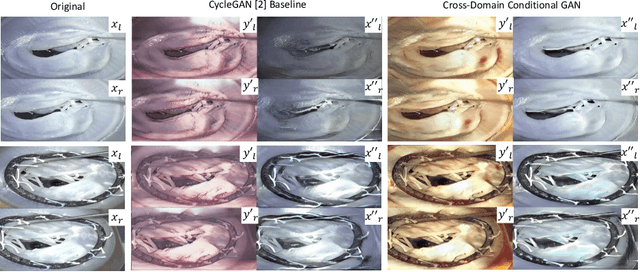
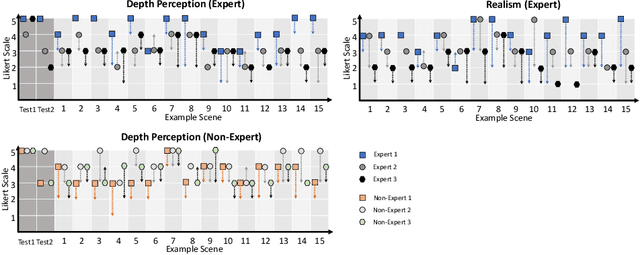
Abstract:Phantoms for surgical training are able to mimic cutting and suturing properties and patient-individual shape of organs, but lack a realistic visual appearance that captures the heterogeneity of surgical scenes. In order to overcome this in endoscopic approaches, hyperrealistic concepts have been proposed to be used in an augmented reality-setting, which are based on deep image-to-image transformation methods. Such concepts are able to generate realistic representations of phantoms learned from real intraoperative endoscopic sequences. Conditioned on frames from the surgical training process, the learned models are able to generate impressive results by transforming unrealistic parts of the image (e.g.\ the uniform phantom texture is replaced by the more heterogeneous texture of the tissue). Image-to-image synthesis usually learns a mapping $G:X~\to~Y$ such that the distribution of images from $G(X)$ is indistinguishable from the distribution $Y$. However, it does not necessarily force the generated images to be consistent and without artifacts. In the endoscopic image domain this can affect depth cues and stereo consistency of a stereo image pair, which ultimately impairs surgical vision. We propose a cross-domain conditional generative adversarial network approach (GAN) that aims to generate more consistent stereo pairs. The results show substantial improvements in depth perception and realism evaluated by 3 domain experts and 3 medical students on a 3D monitor over the baseline method. In 84 of 90 instances our proposed method was preferred or rated equal to the baseline.
Automatic Cardiac Disease Assessment on cine-MRI via Time-Series Segmentation and Domain Specific Features
Jan 25, 2018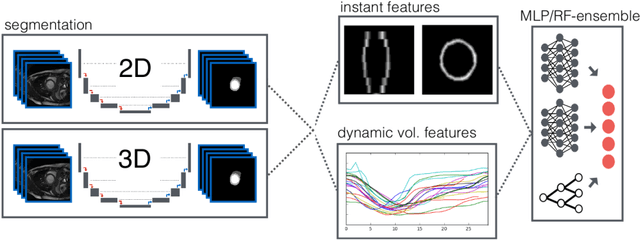
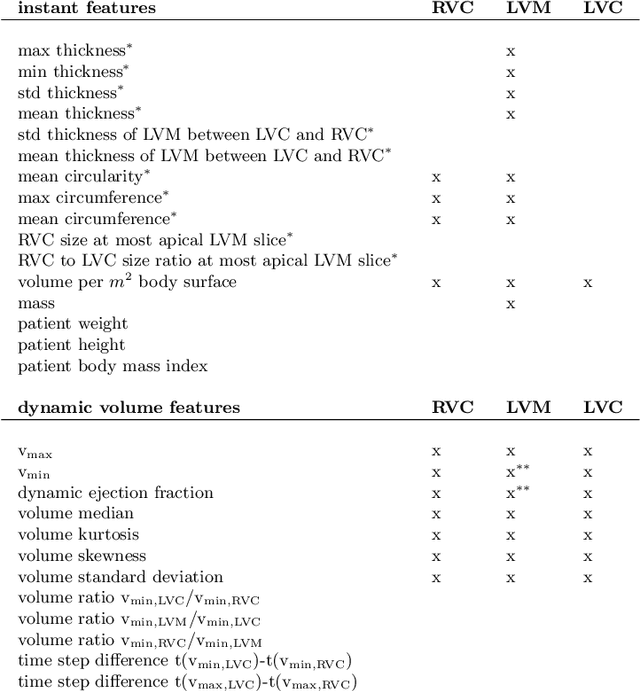
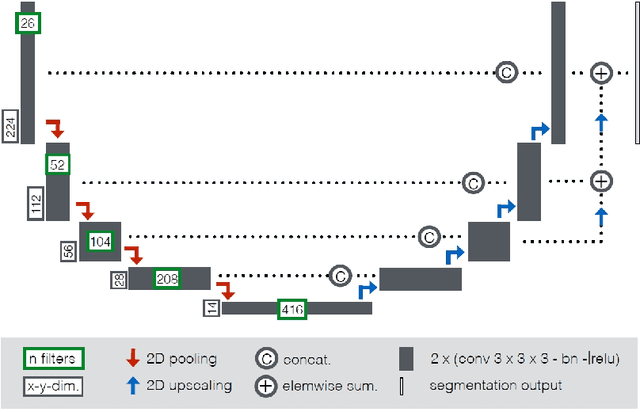
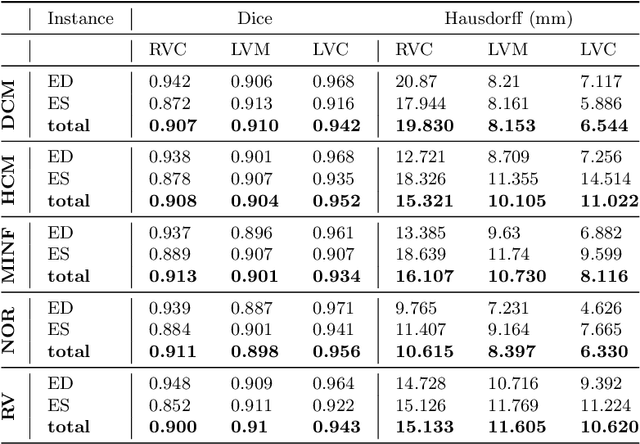
Abstract:Cardiac magnetic resonance imaging improves on diagnosis of cardiovascular diseases by providing images at high spatiotemporal resolution. Manual evaluation of these time-series, however, is expensive and prone to biased and non-reproducible outcomes. In this paper, we present a method that addresses named limitations by integrating segmentation and disease classification into a fully automatic processing pipeline. We use an ensemble of UNet inspired architectures for segmentation of cardiac structures such as the left and right ventricular cavity (LVC, RVC) and the left ventricular myocardium (LVM) on each time instance of the cardiac cycle. For the classification task, information is extracted from the segmented time-series in form of comprehensive features handcrafted to reflect diagnostic clinical procedures. Based on these features we train an ensemble of heavily regularized multilayer perceptrons (MLP) and a random forest classifier to predict the pathologic target class. We evaluated our method on the ACDC dataset (4 pathology groups, 1 healthy group) and achieve dice scores of 0.945 (LVC), 0.908 (RVC) and 0.905 (LVM) in a cross-validation over the training set (100 cases) and 0.950 (LVC), 0.923 (RVC) and 0.911 (LVM) on the test set (50 cases). We report a classification accuracy of 94% on a training set cross-validation and 92% on the test set. Our results underpin the potential of machine learning methods for accurate, fast and reproducible segmentation and computer-assisted diagnosis (CAD).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge