Philipp Beerbaum
Artificial Intelligence in Pediatric Echocardiography: Exploring Challenges, Opportunities, and Clinical Applications with Explainable AI and Federated Learning
Nov 15, 2024


Abstract:Pediatric heart diseases present a broad spectrum of congenital and acquired diseases. More complex congenital malformations require a differentiated and multimodal decision-making process, usually including echocardiography as a central imaging method. Artificial intelligence (AI) offers considerable promise for clinicians by facilitating automated interpretation of pediatric echocardiography data. However, adapting AI technologies for pediatric echocardiography analysis has challenges such as limited public data availability, data privacy, and AI model transparency. Recently, researchers have focused on disruptive technologies, such as federated learning (FL) and explainable AI (XAI), to improve automatic diagnostic and decision support workflows. This study offers a comprehensive overview of the limitations and opportunities of AI in pediatric echocardiography, emphasizing the synergistic workflow and role of XAI and FL, identifying research gaps, and exploring potential future developments. Additionally, three relevant clinical use cases demonstrate the functionality of XAI and FL with a focus on (i) view recognition, (ii) disease classification, (iii) segmentation of cardiac structures, and (iv) quantitative assessment of cardiac function.
MedDoc-Bot: A Chat Tool for Comparative Analysis of Large Language Models in the Context of the Pediatric Hypertension Guideline
May 06, 2024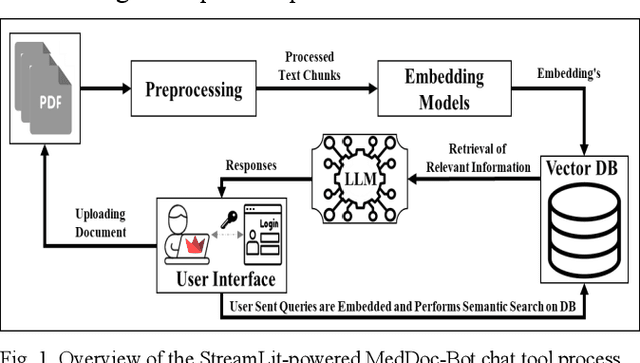
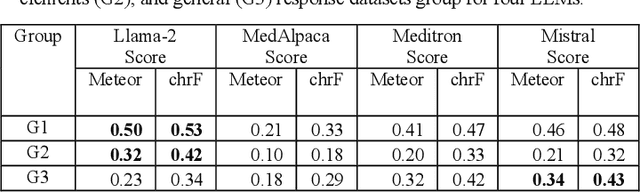
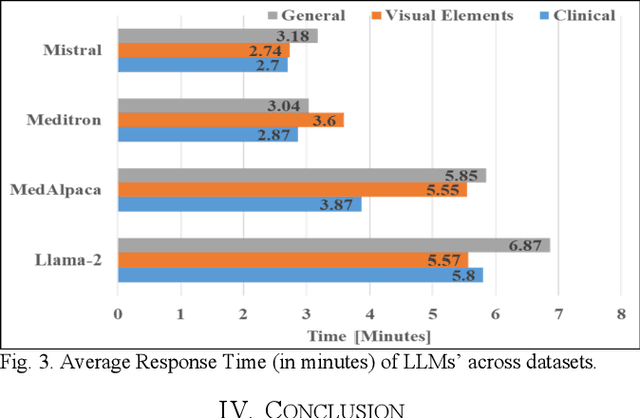
Abstract:This research focuses on evaluating the non-commercial open-source large language models (LLMs) Meditron, MedAlpaca, Mistral, and Llama-2 for their efficacy in interpreting medical guidelines saved in PDF format. As a specific test scenario, we applied these models to the guidelines for hypertension in children and adolescents provided by the European Society of Cardiology (ESC). Leveraging Streamlit, a Python library, we developed a user-friendly medical document chatbot tool (MedDoc-Bot). This tool enables authorized users to upload PDF files and pose questions, generating interpretive responses from four locally stored LLMs. A pediatric expert provides a benchmark for evaluation by formulating questions and responses extracted from the ESC guidelines. The expert rates the model-generated responses based on their fidelity and relevance. Additionally, we evaluated the METEOR and chrF metric scores to assess the similarity of model responses to reference answers. Our study found that Llama-2 and Mistral performed well in metrics evaluation. However, Llama-2 was slower when dealing with text and tabular data. In our human evaluation, we observed that responses created by Mistral, Meditron, and Llama-2 exhibited reasonable fidelity and relevance. This study provides valuable insights into the strengths and limitations of LLMs for future developments in medical document interpretation. Open-Source Code: https://github.com/yaseen28/MedDoc-Bot
How well do U-Net-based segmentation trained on adult cardiac magnetic resonance imaging data generalise to rare congenital heart diseases for surgical planning?
Feb 10, 2020



Abstract:Planning the optimal time of intervention for pulmonary valve replacement surgery in patients with the congenital heart disease Tetralogy of Fallot (TOF) is mainly based on ventricular volume and function according to current guidelines. Both of these two biomarkers are most reliably assessed by segmentation of 3D cardiac magnetic resonance (CMR) images. In several grand challenges in the last years, U-Net architectures have shown impressive results on the provided data. However, in clinical practice, data sets are more diverse considering individual pathologies and image properties derived from different scanner properties. Additionally, specific training data for complex rare diseases like TOF is scarce. For this work, 1) we assessed the accuracy gap when using a publicly available labelled data set (the Automatic Cardiac Diagnosis Challenge (ACDC) data set) for training and subsequent applying it to CMR data of TOF patients and vice versa and 2) whether we can achieve similar results when applying the model to a more heterogeneous data base. Multiple deep learning models were trained with four-fold cross validation. Afterwards they were evaluated on the respective unseen CMR images from the other collection. Our results confirm that current deep learning models can achieve excellent results (left ventricle dice of $0.951\pm{0.003}$/$0.941\pm{0.007}$ train/validation) within a single data collection. But once they are applied to other pathologies, it becomes apparent how much they overfit to the training pathologies (dice score drops between $0.072\pm{0.001}$ for the left and $0.165\pm{0.001}$ for the right ventricle).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge